Minor prion substrains overcome transmission barriers
- PMID: 39440977
- PMCID: PMC11559082
- DOI: 10.1128/mbio.02721-24
Minor prion substrains overcome transmission barriers
Abstract
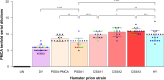
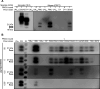
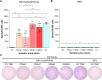
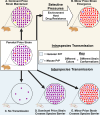
Mammalian prion diseases are infectious neurodegenerative diseases caused by the self-templating form of the prion protein PrPSc. Much evidence supports the hypothesis that prions exist as a mixture of a dominant strain and minor prion strains. While it is known that prions can infect new species, the relative contribution of the dominant prion strain and minor strains in crossing the species barrier is unknown. We previously identified minor prion strains from a biologically cloned drowsy (DY) strain of hamster-adapted transmissible mink encephalopathy (TME). Here we show that these minor prion strains have increased infection efficiency to rabbit kidney epithelial cells that express hamster PrPC compared to the dominant strain DY TME. Using protein misfolding cyclic amplification (PMCA), we found that the dominant strain DY TME failed to convert mouse PrPC to PrPSc, even after several serial passages. In contrast, the minor prion strains isolated from biologically cloned DY TME robustly converted mouse PrPC to PrPSc in the first round of PMCA. This observation indicates that minor prion strains from the mutant spectra contribute to crossing the species barrier. Additionally, we found that the PMCA conversion efficiency for the minor prion strains tested was significantly different from each other and from the short-incubation period prion strain HY TME. This suggests that minor strain diversity may be greater than previously anticipated. These observations further expand our understanding of the mechanisms underlying the species barrier effect and has implications for assessing the zoonotic potential of prions.
Importance: Prions from cattle with bovine spongiform encephalopathy have transmitted to humans, whereas scrapie from sheep and goats likely has not, suggesting that some prions can cross species barriers more easily than others. Prions are composed of a dominant strain and minor strains, and the contribution of each population to adapt to new replicative environments is unknown. Recently, minor prion strains were isolated from the biologically cloned prion strain DY TME, and these minor prion strains differed in properties from the dominant prion strain, DY TME. Here we found that these minor prion strains also differed in conversion efficiency and host range compared to the dominant strain DY TME. These novel findings provide evidence that minor prion strains contribute to interspecies transmission, underscoring the significance of minor strain components in important biological processes.
Keywords: interspecies transmission; prion disease; prion strain.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
The effectiveness of abstinence-based and harm reduction-based interventions in reducing problematic substance use in adults who are experiencing homelessness in high income countries: A systematic review and meta-analysis: A systematic review.Campbell Syst Rev. 2024 Apr 21;20(2):e1396. doi: 10.1002/cl2.1396. eCollection 2024 Jun. Campbell Syst Rev. 2024. PMID: 38645303 Free PMC article. Review.
-
Screening of Anti-Prion Compounds Using the Protein Misfolding Cyclic Amplification Technology.Biomolecules. 2024 Sep 4;14(9):1113. doi: 10.3390/biom14091113. Biomolecules. 2024. PMID: 39334879 Free PMC article.
-
Healthcare workers' informal uses of mobile phones and other mobile devices to support their work: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2024 Aug 27;8(8):CD015705. doi: 10.1002/14651858.CD015705.pub2. Cochrane Database Syst Rev. 2024. PMID: 39189465 Free PMC article.
-
Using Experience Sampling Methodology to Capture Disclosure Opportunities for Autistic Adults.Autism Adulthood. 2023 Dec 1;5(4):389-400. doi: 10.1089/aut.2022.0090. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116059 Free PMC article.
References
-
- Kong Q, Bessen RA. 2017. Prion diseases, p 517–531. In Ikezu T, Gendelman HE (ed), Neuroimmune pharmacology. Springer international publishing, Switzerland.
-
- Ironside JW, Ritchie DL, Head MW. 2018. Prion diseases, p 393–403. In Kovacs GG, Alafuzoff I (ed), Neuropathology. Elsevier B.V.
-
- Orge L, Lima C, Machado C, Tavares P, Mendonça P, Carvalho P, Silva J, Pinto M de L, Bastos E, Pereira JC, Gonçalves-Anjo N, Gama A, Esteves A, Alves A, Matos AC, Seixas F, Silva F, Pires I, Figueira L, Vieira-Pinto M, Sargo R, Pires MDA. 2021. Neuropathology of animal prion diseases. Biomolecules 11:466. doi:10.3390/biom11030466 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- R01NS133050/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01NS103763/HHS | NIH | National Institute of Neurological Disorders and Stroke (NINDS)
- R01 NS103763/NS/NINDS NIH HHS/United States
- P01 AI077774/AI/NIAID NIH HHS/United States
- R01 NS133050/NS/NINDS NIH HHS/United States
LinkOut - more resources
Full Text Sources
Research Materials
