Inflammatory profile of eosinophils in asthma-COPD overlap and eosinophilic COPD: a multi-omics study
- PMID: 39439801
- PMCID: PMC11493663
- DOI: 10.3389/fimmu.2024.1445769
Inflammatory profile of eosinophils in asthma-COPD overlap and eosinophilic COPD: a multi-omics study
Abstract
Introduction: Elevated blood eosinophil levels in patients with chronic obstructive pulmonary disease (COPD) with or without asthma are linked to increased exacerbations and the effectiveness of inhaled corticosteroid treatment. This study aimed to delineate the inflammatory cellular properties of eosinophils in patients with asthma-COPD overlap (ACO) and eosinophilic COPD (eCOPD).
Methods: Eosinophils were isolated from the peripheral blood of healthy volunteers, patients with non-eCOPD, and those with ACO/eCOPD. Multi-omics analysis involving transcriptomics, proteomics, and lipidomics was performed, followed by bioinformatic data analyses. In vitro experiments using eosinophils from healthy volunteers were conducted to investigate the molecular mechanisms underlying cellular alterations in eosinophils.
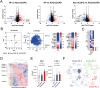
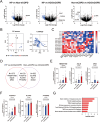
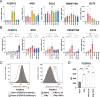
Results: Proteomics and transcriptomics analyses revealed cellular characteristics in overall COPD patients represented by viral infection (elevated expression of sterol regulatory element-binding protein-1) and inflammatory responses (elevated levels of IL1 receptor-like 1, Fc epsilon receptor Ig, and transmembrane protein 176B). Cholesterol metabolism enzymes were identified as ACO/eCOPD-related factors. Gene Ontology and pathway enrichment analyses demonstrated the key roles of antiviral responses, cholesterol metabolism, and inflammatory molecules-related signaling pathways in ACO/eCOPD. Lipidomics showed the impaired synthesis of cyclooxygenase-derived mediators including prostaglandin E2 (PGE2) in ACO/eCOPD. In vitro assessment confirmed that IL-33 or TNF-α stimulation combined with IL-5 and IFN-γ stimulation induced cellular signatures in eosinophils in ACO/eCOPD. Atorvastatin, dexamethasone, and PGE2 differentially modulated these inflammatory changes.
Discussion: ACO/eCOPD is associated with viral infection and an inflammatory milieu. Therapeutic strategies using statins and inhaled corticosteroids are recommended to control these pathogenic changes.
Keywords: IL-33; asthma-COPD overlap; chronic obstructive pulmonary disease; eosinophil; interferon-γ; multi-omics; statin; tumor necrosis factor-α.
Copyright © 2024 Sunata, Miyata, Kawashima, Konno, Ishikawa, Hasegawa, Onozato, Otsu, Matsuyama, Sasaki, Okuzumi, Mochimaru, Masaki, Kabata, Chubachi, Arita and Fukunaga.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures


Similar articles
-
Disease burden of eosinophilic airway disease: Comparing severe asthma, COPD and asthma-COPD overlap.Respirology. 2021 Jan;26(1):52-61. doi: 10.1111/resp.13841. Epub 2020 May 19. Respirology. 2021. PMID: 32428971
-
Modulation of blood inflammatory markers by benralizumab in patients with eosinophilic airway diseases.Respir Res. 2019 Jan 18;20(1):14. doi: 10.1186/s12931-018-0968-8. Respir Res. 2019. PMID: 30658649 Free PMC article. Clinical Trial.
-
Metabolomic fingerprinting and systemic inflammatory profiling of asthma COPD overlap (ACO).Respir Res. 2020 May 24;21(1):126. doi: 10.1186/s12931-020-01390-4. Respir Res. 2020. PMID: 32448302 Free PMC article.
-
Role of eosinophils in airway inflammation of chronic obstructive pulmonary disease.Int J Chron Obstruct Pulmon Dis. 2018 Jan 17;13:335-349. doi: 10.2147/COPD.S152291. eCollection 2018. Int J Chron Obstruct Pulmon Dis. 2018. PMID: 29403271 Free PMC article. Review.
-
The pharmacological management of asthma-chronic obstructive pulmonary disease overlap syndrome (ACOS).Expert Opin Pharmacother. 2020 Feb;21(2):213-231. doi: 10.1080/14656566.2019.1701656. Expert Opin Pharmacother. 2020. PMID: 31955671 Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

