Oncometabolites at the crossroads of genetic, epigenetic and ecological alterations in cancer
- PMID: 39438765
- PMCID: PMC11618380
- DOI: 10.1038/s41418-024-01402-6
Oncometabolites at the crossroads of genetic, epigenetic and ecological alterations in cancer
Abstract
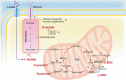
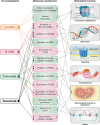
By the time a tumor reaches clinical detectability, it contains around 108-109 cells. However, during tumor formation, significant cell loss occurs due to cell death. In some estimates, it could take up to a thousand cell generations, over a ~ 20-year life-span of a tumor, to reach clinical detectability, which would correspond to a "theoretical" generation of ~1030 cells. These rough calculations indicate that cancers are under negative selection. The fact that they thrive implies that they "evolve", and that their evolutionary trajectories are shaped by the pressure of the environment. Evolvability of a cancer is a function of its heterogeneity, which could be at the genetic, epigenetic, and ecological/microenvironmental levels [1]. These principles were summarized in a proposed classification in which Evo (evolutionary) and Eco (ecological) indexes are used to label cancers [1]. The Evo index addresses cancer cell-autonomous heterogeneity (genetic/epigenetic). The Eco index describes the ecological landscape (non-cell-autonomous) in terms of hazards to cancer survival and resources available. The reciprocal influence of Evo and Eco components is critical, as it can trigger self-sustaining loops that shape cancer evolvability [2]. Among the various hallmarks of cancer [3], metabolic alterations appear unique in that they intersect with both Evo and Eco components. This is partly because altered metabolism leads to the accumulation of oncometabolites. These oncometabolites have traditionally been viewed as mediators of non-cell-autonomous alterations in the cancer microenvironment. However, they are now increasingly recognized as inducers of genetic and epigenetic modifications. Thus, oncometabolites are uniquely positioned at the crossroads of genetic, epigenetic and ecological alterations in cancer. In this review, the mechanisms of action of oncometabolites will be summarized, together with their roles in the Evo and Eco phenotypic components of cancer evolvability. An evolutionary perspective of the impact of oncometabolites on the natural history of cancer will be presented.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The author declares no competing of interest.
Figures


Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Far Posterior Approach for Rib Fracture Fixation: Surgical Technique and Tips.JBJS Essent Surg Tech. 2024 Dec 6;14(4):e23.00094. doi: 10.2106/JBJS.ST.23.00094. eCollection 2024 Oct-Dec. JBJS Essent Surg Tech. 2024. PMID: 39650795 Free PMC article.
-
Defining the optimum strategy for identifying adults and children with coeliac disease: systematic review and economic modelling.Health Technol Assess. 2022 Oct;26(44):1-310. doi: 10.3310/ZUCE8371. Health Technol Assess. 2022. PMID: 36321689 Free PMC article.
-
"I've Spent My Whole Life Striving to Be Normal": Internalized Stigma and Perceived Impact of Diagnosis in Autistic Adults.Autism Adulthood. 2023 Dec 1;5(4):423-436. doi: 10.1089/aut.2022.0066. Epub 2023 Dec 12. Autism Adulthood. 2023. PMID: 38116050 Free PMC article.
-
Trends in Surgical and Nonsurgical Aesthetic Procedures: A 14-Year Analysis of the International Society of Aesthetic Plastic Surgery-ISAPS.Aesthetic Plast Surg. 2024 Oct;48(20):4217-4227. doi: 10.1007/s00266-024-04260-2. Epub 2024 Aug 5. Aesthetic Plast Surg. 2024. PMID: 39103642 Review.
References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

