Elucidating gastric cancer mechanisms and therapeutic potential of Adociaquinone A targeting EGFR: A genomic analysis and Computer Aided Drug Design (CADD) approach
- PMID: 39434198
- PMCID: PMC11493557
- DOI: 10.1111/jcmm.70133
Elucidating gastric cancer mechanisms and therapeutic potential of Adociaquinone A targeting EGFR: A genomic analysis and Computer Aided Drug Design (CADD) approach
Abstract
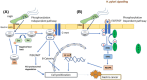
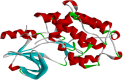
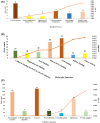
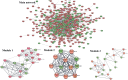
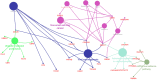
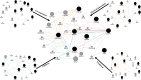
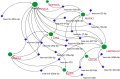
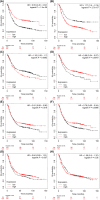
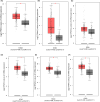
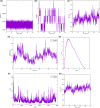
Gastric cancer predominantly adenocarcinoma, accounts for over 85% of gastric cancer diagnoses. Current therapeutic options are limited, necessitating the discovery of novel drug targets and effective treatments. The Affymetrix gene expression microarray dataset (GSE64951) was retrieved from NCBI-GEO data normalization and DEGs identification was done by using R-Bioconductor package. Gene Ontology (GO) analysis of DEGs was performed using DAVID. The protein-protein interaction network was constructed by STRING database plugin in Cytoscape. Subclusters/modules of important interacting genes in main network were extracted by using MCODE. The hub genes from in the network were identified by using Cytohubba. The miRNet tool built a hub gene/mRNA-miRNA network and Kaplan-Meier-Plotter conducted survival analysis. AutoDock Vina and GROMACS MD simulations were used for docking and stability analysis of marine compounds against the 5CNN protein. Total 734 DEGs (507 up-regulated and 228 down-regulated) were identified. Differentially expressed genes (DEGs) were enriched in processes like cell-cell adhesion and ATP binding. Eight hub genes (EGFR, HSPA90AA1, MAPK1, HSPA4, PPP2CA, CDKN2A, CDC20, and ATM) were selected for further analysis. A total of 23 miRNAs associated with hub genes were identified, with 12 of them targeting PPP2CA. EGFR displayed the highest expression and hazard rate in survival analyses. The kinase domain of EGFR (PDBID: 5CNN) was chosen as the drug target. Adociaquinone A from Petrosia alfiani, docked with 5CNN, showed the lowest binding energy with stable interactions across a 50 ns MD simulation, highlighting its potential as a lead molecule against EGFR. This study has identified crucial DEGs and hub genes in gastric cancer, proposing novel therapeutic targets. Specifically, Adociaquinone A demonstrates promising potential as a bioactive drug against EGFR in gastric cancer, warranting further investigation. The predicted miRNA against the hub gene/proteins can also be used as potential therapeutic targets.
Keywords: CMNPD; hub genes; miRNA; molecular docking; survival analysis.
© 2024 The Author(s). Journal of Cellular and Molecular Medicine published by Foundation for Cellular and Molecular Medicine and John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
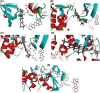

Similar articles
-
Screening and Survival Analysis of Hub Genes in Gastric Cancer Based on Bioinformatics.J Comput Biol. 2019 Nov;26(11):1316-1325. doi: 10.1089/cmb.2019.0119. Epub 2019 Jun 24. J Comput Biol. 2019. PMID: 31233344
-
Integrated bioinformatics analysis for the screening of hub genes and therapeutic drugs in ovarian cancer.J Ovarian Res. 2020 Jan 27;13(1):10. doi: 10.1186/s13048-020-0613-2. J Ovarian Res. 2020. PMID: 31987036 Free PMC article.
-
Integrated Bioinformatics Analysis for the Screening of Hub Genes and Therapeutic Drugs in Hepatocellular Carcinoma.Curr Pharm Biotechnol. 2023;24(8):1035-1058. doi: 10.2174/1389201023666220628113452. Curr Pharm Biotechnol. 2023. PMID: 35762549
-
Identification of hub genes with prognostic values in gastric cancer by bioinformatics analysis.World J Surg Oncol. 2018 Jun 19;16(1):114. doi: 10.1186/s12957-018-1409-3. World J Surg Oncol. 2018. PMID: 29921304 Free PMC article.
-
Identification of potential miRNA-mRNA regulatory network contributing to pathogenesis of HBV-related HCC.J Transl Med. 2019 Jan 3;17(1):7. doi: 10.1186/s12967-018-1761-7. J Transl Med. 2019. PMID: 30602391 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

