Mitochondrial mechanotransduction through MIEF1 coordinates the nuclear response to forces
- PMID: 39433949
- PMCID: PMC11628398
- DOI: 10.1038/s41556-024-01527-3
Mitochondrial mechanotransduction through MIEF1 coordinates the nuclear response to forces
Abstract
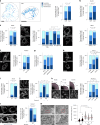
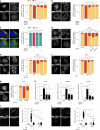
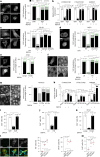
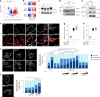
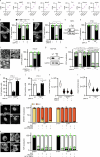
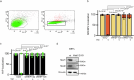
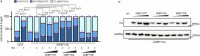
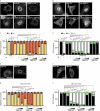
Tissue-scale architecture and mechanical properties instruct cell behaviour under physiological and diseased conditions, but our understanding of the underlying mechanisms remains fragmentary. Here we show that extracellular matrix stiffness, spatial confinements and applied forces, including stretching of mouse skin, regulate mitochondrial dynamics. Actomyosin tension promotes the phosphorylation of mitochondrial elongation factor 1 (MIEF1), limiting the recruitment of dynamin-related protein 1 (DRP1) at mitochondria, as well as peri-mitochondrial F-actin formation and mitochondrial fission. Strikingly, mitochondrial fission is also a general mechanotransduction mechanism. Indeed, we found that DRP1- and MIEF1/2-dependent fission is required and sufficient to regulate three transcription factors of broad relevance-YAP/TAZ, SREBP1/2 and NRF2-to control cell proliferation, lipogenesis, antioxidant metabolism, chemotherapy resistance and adipocyte differentiation in response to mechanical cues. This extends to the mouse liver, where DRP1 regulates hepatocyte proliferation and identity-hallmark YAP-dependent phenotypes. We propose that mitochondria fulfil a unifying signalling function by which the mechanical tissue microenvironment coordinates complementary cell functions.
© 2024. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures









Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
BMAL1 and CLOCK proteins exhibit differential association with mitochondrial dynamics, protein synthesis pathways and muscle strength in human muscle.J Physiol. 2024 Dec;602(23):6403-6415. doi: 10.1113/JP285955. Epub 2024 Jun 26. J Physiol. 2024. PMID: 38922907
-
PIEZO1 mediates matrix stiffness-induced tumor progression in kidney renal clear cell carcinoma by activating the Ca2+/Calpain/YAP pathway.Biochim Biophys Acta Mol Cell Res. 2025 Jan;1872(1):119871. doi: 10.1016/j.bbamcr.2024.119871. Epub 2024 Oct 28. Biochim Biophys Acta Mol Cell Res. 2025. PMID: 39490703
-
A role for septin 2 in Drp1-mediated mitochondrial fission.EMBO Rep. 2016 Jun;17(6):858-73. doi: 10.15252/embr.201541612. Epub 2016 May 23. EMBO Rep. 2016. PMID: 27215606 Free PMC article.
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
References
-
- Mohammadi, H. & Sahai, E. Mechanisms and impact of altered tumour mechanics. Nat. Cell Biol.20, 766–774 (2018). - PubMed
MeSH terms
Substances
Grants and funding
- 21392/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- 28940/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- MFAG 27453/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- MFAG 24856/Associazione Italiana per la Ricerca sul Cancro (Italian Association for Cancer Research)
- 2022T9RM8A/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- B73C22001250006 MechanoMet/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- P2022CE7SP/Ministero dell'Istruzione, dell'Università e della Ricerca (Ministry of Education, University and Research)
- Eccellenza 2023 MINDED/Fondazione Cassa di Risparmio di Padova e Rovigo (Foundation Cariparo)
- 2022/Fondazione Umberto Veronesi (Umberto Veronesi Foundation)
- 2023/Fondazione Umberto Veronesi (Umberto Veronesi Foundation)
- NNF21CC0073729/Novo Nordisk Fonden (Novo Nordisk Foundation)
- 205321_188828/Schweizerischer Nationalfonds zur Förderung der Wissenschaftlichen Forschung (Swiss National Science Foundation)
- TMPGMFU22TT/Fondazione Telethon (Telethon Foundation)
- MetEpiStem/EC | EC Seventh Framework Programm | FP7 Ideas: European Research Council (FP7-IDEAS-ERC - Specific Programme: "Ideas" Implementing the Seventh Framework Programme of the European Community for Research, Technological Development and Demonstration Activities (2007 to 2013))
- 21-0156/AICR_/Worldwide Cancer Research/United Kingdom
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

