Lytic promoter activity during herpes simplex virus latency is dependent on genome location
- PMID: 39431845
- PMCID: PMC11575402
- DOI: 10.1128/jvi.01258-24
Lytic promoter activity during herpes simplex virus latency is dependent on genome location
Abstract
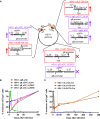
Herpes simplex virus 1 (HSV-1) is a significant pathogen that establishes lifelong latent infections with intermittent episodes of resumed disease. In mouse models of HSV infection, sporadic low-level lytic gene expression has been detected during latency in the absence of reactivation events that lead to production of new viruses. This viral activity during latency has been reported using a sensitive Cre-marking model for several lytic gene promoters placed in one location in the HSV-1 genome. Here, we extend these findings in the same model by examining first, the activity of an ectopic lytic gene promoter in several places in the genome and second, whether any promoters might be active in their natural context. We found that Cre expression was detected during latency from ectopic and native promoters, but only in locations near the ends of the unique long genome segment. This location is significant because it is in close proximity to the region from which latency-associated transcripts (LATs) are derived. These results show that native HSV-1 lytic gene promoters can produce protein products during latency, but that this activity is only detectable when they are located close to the LAT locus.IMPORTANCEHSV is a significant human pathogen and the best studied model of mammalian virus latency. Traditionally, the active (lytic) and inactive (latent) phases of infection were considered to be distinct, but the notion of latency being entirely quiescent is evolving due to the detection of some lytic gene expression during latency. Here, we add to this literature by finding that the activity can be found for native lytic gene promoters as well as for constructs placed ectopically in the HSV genome. However, this activity was only detectable when these promoters were located close by a region known to be transcriptionally active during latency. These data have implications for our understanding of HSV gene regulation during latency and the extent to which transcriptionally active regions are insulated from adjacent parts of the viral genome.
Keywords: gene expression; herpes simplex virus; latency.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
HSV-1 LAT Promoter Deletion Viruses Exhibit Strain-Specific and LAT-Dependent Epigenetic Regulation of Latent Viral Genomes in Human Neurons.J Virol. 2023 Feb 28;97(2):e0193522. doi: 10.1128/jvi.01935-22. Epub 2023 Feb 1. J Virol. 2023. PMID: 36722973 Free PMC article.
-
A protein encoded by the herpes simplex virus (HSV) type 1 2-kilobase latency-associated transcript is phosphorylated, localized to the nucleus, and overcomes the repression of expression from exogenous promoters when inserted into the quiescent HSV genome.J Virol. 2002 Apr;76(8):4056-67. doi: 10.1128/jvi.76.8.4056-4067.2002. J Virol. 2002. PMID: 11907244 Free PMC article.
-
Two herpes simplex virus type 1 latency-active promoters differ in their contributions to latency-associated transcript expression during lytic and latent infections.J Virol. 1995 Dec;69(12):7899-908. doi: 10.1128/JVI.69.12.7899-7908.1995. J Virol. 1995. PMID: 7494302 Free PMC article.
-
Molecular circuitry regulating herpes simplex virus type 1 latency in neurons.J Neurovirol. 2000 Feb;6(1):6-24. doi: 10.3109/13550280009006378. J Neurovirol. 2000. PMID: 10786993 Review.
-
A comparison of herpes simplex virus type 1 and varicella-zoster virus latency and reactivation.J Gen Virol. 2015 Jul;96(Pt 7):1581-602. doi: 10.1099/vir.0.000128. Epub 2015 Mar 20. J Gen Virol. 2015. PMID: 25794504 Free PMC article. Review.
References
-
- Conley AJ, Knipe DM, Jones PC, Roizman B. 1981. Molecular genetics of herpes simplex virus. VII. Characterization of a temperature-sensitive mutant produced by in vitro mutagenesis and defective in DNA synthesis and accumulation of gamma polypeptides. J Virol 37:191–206. doi:10.1128/JVI.37.1.191-206.1981 - DOI - PMC - PubMed
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

