Molecular Dynamics Simulations Reveal How Competing Protein-Surface Interactions for Glycine, Citrate, and Water Modulate Stability in Antibody Fragment Formulations
- PMID: 39431440
- PMCID: PMC11539065
- DOI: 10.1021/acs.molpharmaceut.4c00332
Molecular Dynamics Simulations Reveal How Competing Protein-Surface Interactions for Glycine, Citrate, and Water Modulate Stability in Antibody Fragment Formulations
Abstract
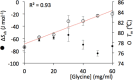
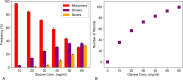
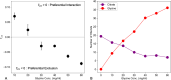
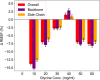
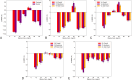
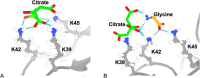
The design of stable formulations remains a major challenge for protein therapeutics, particularly the need to minimize aggregation. Experimental formulation screens are typically based on thermal transition midpoints (Tm), and forced degradation studies at elevated temperatures. Both approaches give limited predictions of long-term storage stability, particularly at low temperatures. Better understanding of the mechanisms of action for formulation of excipients and buffers could lead to improved strategies for formulation design. Here, we identified a complex impact of glycine concentration on the experimentally determined stability of an antibody Fab fragment and then used molecular dynamics simulations to reveal mechanisms that underpin these complex behaviors. Tm values increased monotonically with glycine concentration, but associated ΔSvh measurements revealed more complex changes in the native ensemble dynamics, which reached a maximum at 30 mg/mL. The aggregation kinetics at 65 °C were similar at 0 and 20 mg/mL glycine, but then significantly slower at 50 mg/mL. These complex behaviors indicated changes in the dominant stabilizing mechanisms as the glycine concentration was increased. MD revealed a complex balance of glycine self-interaction, and differentially preferred interactions of glycine with the Fab as it displaced hydration-shell water, and surface-bound water and citrate buffer molecules. As a result, glycine binding to the Fab surface had different effects at different concentrations, and led from preferential interactions at low concentrations to preferential exclusion at higher concentrations. During preferential interaction, glycine displaced water from the Fab hydration shell, and a small number of water and citrate molecules from the Fab surface, which reduced the protein dynamics as measured by root-mean-square fluctuation (RMSF) on the short time scales of MD. By contrast, the native ensemble dynamics increased according to ΔSvh, suggesting increased conformational changes on longer time scales. The aggregation kinetics did not change at low glycine concentrations, and so the opposing dynamics effects either canceled out or were not directly relevant to aggregation. During preferential exclusion at higher glycine concentrations, glycine could only bind to the Fab surface through the displacement of citrate buffer molecules already favorably bound on the Fab surface. Displacement of citrate increased the flexibility (RMSF) of the Fab, as glycine formed fewer bridging hydrogen bonds to the Fab surface. Overall, the slowing of aggregation kinetics coincided with reduced flexibility in the Fab ensemble at the very highest glycine concentrations, as determined by both RMSF and ΔSvh, and occurred at a point where glycine binding displaced neither water nor citrate. These final interactions with the Fab surface were driven by mass action and were the least favorable, leading to a macromolecular crowding effect under the regime of preferential exclusion that stabilized the dynamics of Fab.
Keywords: Fab; aggregation; enthalpy change; formulation; melting temperature; preferential interaction; stability.
Conflict of interest statement
The authors declare no competing financial interest.
Figures
Similar articles
-
Computational Design To Reduce Conformational Flexibility and Aggregation Rates of an Antibody Fab Fragment.Mol Pharm. 2018 Aug 6;15(8):3079-3092. doi: 10.1021/acs.molpharmaceut.8b00186. Epub 2018 Jun 22. Mol Pharm. 2018. PMID: 29897777
-
Enhanced Thermal Stability and Reduced Aggregation in an Antibody Fab Fragment at Elevated Concentrations.Mol Pharm. 2023 May 1;20(5):2650-2661. doi: 10.1021/acs.molpharmaceut.3c00081. Epub 2023 Apr 11. Mol Pharm. 2023. PMID: 37040431 Free PMC article.
-
Macromolecular crowding: chemistry and physics meet biology (Ascona, Switzerland, 10-14 June 2012).Phys Biol. 2013 Aug;10(4):040301. doi: 10.1088/1478-3975/10/4/040301. Epub 2013 Aug 2. Phys Biol. 2013. PMID: 23912807
-
Tm-Values and Unfolded Fraction Can Predict Aggregation Rates for Granulocyte Colony Stimulating Factor Variant Formulations but Not under Predominantly Native Conditions.Mol Pharm. 2018 Jan 2;15(1):256-267. doi: 10.1021/acs.molpharmaceut.7b00876. Epub 2017 Nov 28. Mol Pharm. 2018. PMID: 29141152
-
Computational Characterization of Antibody-Excipient Interactions for Rational Excipient Selection Using the Site Identification by Ligand Competitive Saturation-Biologics Approach.Mol Pharm. 2020 Nov 2;17(11):4323-4333. doi: 10.1021/acs.molpharmaceut.0c00775. Epub 2020 Oct 6. Mol Pharm. 2020. PMID: 32965126 Free PMC article.
References
-
- Jain T.; Sun T.; Durand S.; Hall A.; Houston N. R.; Nett J. H.; Sharkey B.; Bobrowicz B.; Caffry I.; Yu Y.; Cao Y.; Lynaugh H.; Brown M.; Baruah H.; Gray L. T.; Krauland E. M.; Xu Y.; Vásquez M.; Wittrup K. D. Biophysical properties of the clinical-stage antibody landscape. Proc. Natl. Acad. Sci. U.S.A. 2017, 114, 944–949. 10.1073/pnas.1616408114. - DOI - PMC - PubMed
-
- Bailly M.; Mieczkowski C.; Juan V.; Metwally E.; Tomazela D.; Baker J.; Uchida M.; Kofman E.; Raoufi F.; Motlagh S.; Yu Y.; Park J.; Raghava S.; Welsh J.; Rauscher M.; Raghunathan G.; Hsieh M.; Chen Y.-L.; Nguyen H. T.; Nguyen N.; Cipriano D.; Fayadat-Dilman L. Predicting Antibody Developability Profiles Through Early Stage Discovery Screening. MAbs 2020, 12, 174305310.1080/19420862.2020.1743053. - DOI - PMC - PubMed
-
- Dyson M. R.; Masters E.; Pazeraitis D.; Perera R. L.; Syrjanen J. L.; Surade S.; Thorsteinson N.; Parthiban K.; Jones P. C.; Sattar M.; Wozniak-Knopp G.; Rueker F.; Leah R.; McCafferty J. Beyond affinity: selection of antibody variants with optimal biophysical properties and reduced immunogenicity from mammalian display libraries. MAbs 2020, 12, 182933510.1080/19420862.2020.1829335. - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

