Substrate engagement by the intramembrane metalloprotease SpoIVFB
- PMID: 39419996
- PMCID: PMC11486902
- DOI: 10.1038/s41467-024-52634-6
Substrate engagement by the intramembrane metalloprotease SpoIVFB
Abstract
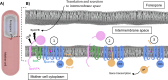
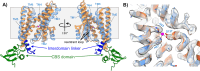
S2P intramembrane metalloproteases regulate diverse signaling pathways across all three domains of life. However, the mechanism by which S2P metalloproteases engage substrates and catalyze peptide hydrolysis within lipid membranes has remained elusive. Here we determine the cryo-EM structure of the S2P family intramembrane metalloprotease SpoIVFB from Bacillus subtilis bound to its native substrate Pro-σK. The structure and accompanying biochemical data demonstrate that SpoIVFB positions Pro-σK at the enzyme active site through a β-sheet augmentation mechanism, and reveal key interactions between Pro-σK and the interdomain linker connecting SpoIVFB transmembrane and CBS domains. The cryo-EM structure and molecular dynamics simulation reveal a plausible path for water to access the membrane-buried active site of SpoIVFB, and suggest a possible role of membrane lipids in facilitating substrate capture. These results provide key insight into how S2P intramembrane metalloproteases capture and position substrates for hydrolytic proteolysis within the hydrophobic interior of a lipid membrane.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Conserved Proline Residues of Bacillus subtilis Intramembrane Metalloprotease SpoIVFB Are Important for Substrate Interaction and Cleavage.J Bacteriol. 2022 Mar 15;204(3):e0038621. doi: 10.1128/JB.00386-21. Epub 2022 Jan 10. J Bacteriol. 2022. PMID: 35007155 Free PMC article.
-
Complex Formed between Intramembrane Metalloprotease SpoIVFB and Its Substrate, Pro-σK.J Biol Chem. 2016 May 6;291(19):10347-62. doi: 10.1074/jbc.M116.715508. Epub 2016 Mar 7. J Biol Chem. 2016. PMID: 26953342 Free PMC article.
-
Interaction of intramembrane metalloprotease SpoIVFB with substrate Pro-σK.Proc Natl Acad Sci U S A. 2017 Dec 12;114(50):E10677-E10686. doi: 10.1073/pnas.1711467114. Epub 2017 Nov 27. Proc Natl Acad Sci U S A. 2017. PMID: 29180425 Free PMC article.
-
Biochemical and structural insights into intramembrane metalloprotease mechanisms.Biochim Biophys Acta. 2013 Dec;1828(12):2873-85. doi: 10.1016/j.bbamem.2013.03.032. Biochim Biophys Acta. 2013. PMID: 24099006 Free PMC article. Review.
-
New insights into S2P signaling cascades: regulation, variation, and conservation.Protein Sci. 2010 Nov;19(11):2015-30. doi: 10.1002/pro.496. Protein Sci. 2010. PMID: 20836086 Free PMC article. Review.
References
-
- Brown, M. S., Ye, J., Rawson, R. B. & Goldstein, J. L. Regulated intramembrane proteolysis: a control mechanism conserved from bacteria to humans. Cell100, 391–398 (2000). - PubMed
-
- Sun, L., Li, X. & Shi, Y. Structural biology of intramembrane proteases: mechanistic insights from rhomboid and S2P to γ-secretase. Curr. Opin. Struct. Biol.37, 97–107 (2016). - PubMed
-
- Beard, H. A., Barniol-Xicota, M., Yang, J. & Verhelst, S. H. L. Discovery of cellular roles of intramembrane proteases. ACS Chem. Biol.14, 2372–2388 (2019). - PubMed
-
- Urban, S. Mechanisms and cellular functions of intramembrane proteases. Biochimica et. biophysica acta1828, 2797–2800 (2013). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

