This is a preprint.
CRISPR tiling deletion screens reveal functional enhancers of neuropsychiatric risk genes and allelic compensation effects (ACE) on transcription
- PMID: 39416108
- PMCID: PMC11483005
- DOI: 10.1101/2024.10.08.616922
CRISPR tiling deletion screens reveal functional enhancers of neuropsychiatric risk genes and allelic compensation effects (ACE) on transcription
Abstract
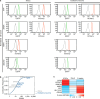
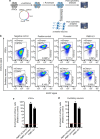
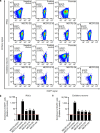
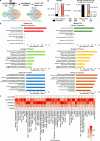
Precise transcriptional regulation is critical for cellular function and development, yet the mechanism of this process remains poorly understood for many genes. To gain a deeper understanding of the regulation of neuropsychiatric disease risk genes, we identified a total of 39 functional enhancers for four dosage-sensitive genes, APP, FMR1, MECP2, and SIN3A, using CRISPR tiling deletion screening in human induced pluripotent stem cell (iPSC)-induced excitatory neurons. We found that enhancer annotation provides potential pathological insights into disease-associated copy number variants. More importantly, we discovered that allelic enhancer deletions at SIN3A could be compensated by increased transcriptional activities from the other intact allele. Such allelic compensation effects (ACE) on transcription is stably maintained during differentiation and, once established, cannot be reversed by ectopic SIN3A expression. Further, ACE at SIN3A occurs through dosage sensing by the promoter. Together, our findings unravel a regulatory compensation mechanism that ensures stable and precise transcriptional output for SIN3A, and potentially other dosage-sensitive genes.
Conflict of interest statement
Competing interests B.R. is a co-founder and consultant of Arima Genomics Inc. and co-founder of Epigenome Technologies. The other authors declare that they have no competing interests.
Figures













Similar articles
-
Enhancers and super-enhancers have an equivalent regulatory role in embryonic stem cells through regulation of single or multiple genes.Genome Res. 2017 Feb;27(2):246-258. doi: 10.1101/gr.210930.116. Epub 2016 Nov 28. Genome Res. 2017. PMID: 27895109 Free PMC article.
-
Functional characterization of QT interval associated SCN5A enhancer variants identify combined additive effects.bioRxiv [Preprint]. 2024 Mar 14:2024.03.11.584440. doi: 10.1101/2024.03.11.584440. bioRxiv. 2024. Update in: HGG Adv. 2024 Sep 30;6(1):100358. doi: 10.1016/j.xhgg.2024.100358 PMID: 38559211 Free PMC article. Updated. Preprint.
-
CRISPR Screening Uncovers a Long-Range Enhancer for ONECUT1 in Pancreatic Differentiation and Links a Diabetes Risk Variant.bioRxiv [Preprint]. 2024 Jun 28:2024.04.26.591412. doi: 10.1101/2024.04.26.591412. bioRxiv. 2024. Update in: Cell Rep. 2024 Aug 27;43(8):114640. doi: 10.1016/j.celrep.2024.114640 PMID: 38746154 Free PMC article. Updated. Preprint.
-
Duplication Versus Deletion Through the Lens of 15q13.3: Clinical and Research Implications of Studying Copy Number Variants Associated with Neuropsychiatric Disorders in Induced Pluripotent Stem Cell-Derived Neurons.Stem Cell Rev Rep. 2023 Apr;19(3):639-650. doi: 10.1007/s12015-022-10475-0. Epub 2022 Nov 12. Stem Cell Rev Rep. 2023. PMID: 36370261 Free PMC article. Review.
-
Hooked Up from a Distance: Charting Genome-Wide Long-Range Interaction Maps in Neural Cells Chromatin to Identify Novel Candidate Genes for Neurodevelopmental Disorders.Int J Mol Sci. 2023 Jan 6;24(2):1164. doi: 10.3390/ijms24021164. Int J Mol Sci. 2023. PMID: 36674677 Free PMC article. Review.
References
-
- Lettice L. A. et al. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12, 1725–1735 (2003). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
