This is a preprint.
map3k1 suppresses terminal differentiation of migratory eye progenitors in planarian regeneration
- PMID: 39416008
- PMCID: PMC11483071
- DOI: 10.1101/2024.10.11.617745
map3k1 suppresses terminal differentiation of migratory eye progenitors in planarian regeneration
Abstract
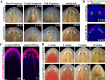
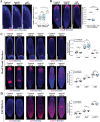
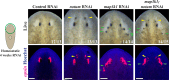
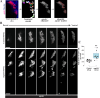
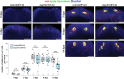
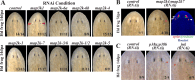
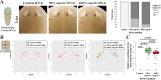
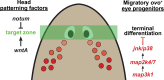
Proper stem cell targeting and differentiation is necessary for regeneration to succeed. In organisms capable of whole body regeneration, considerable progress has been made identifying wound signals initiating this process, but the mechanisms that control the differentiation of progenitors into mature organs are not fully understood. Using the planarian as a model system, we identify a novel function for map3k1, a MAP3K family member possessing both kinase and ubiquitin ligase domains, to negatively regulate terminal differentiation of stem cells during eye regeneration. Inhibition of map3k1 caused the formation of multiple ectopic eyes within the head, but without controlling overall head, brain, or body patterning. By contrast, other known regulators of planarian eye patterning like WntA and notum also regulate head regionalization, suggesting map3k1 acts distinctly. Eye resection and regeneration experiments suggest that unlike Wnt signaling perturbation, map3k1 inhibition did not shift the target destination of eye formation in the animal. Instead, map3k1(RNAi) ectopic eyes emerge in the regions normally occupied by migratory eye progenitors, and the onset of ectopic eyes after map3k1 inhibition coincides with a reduction to eye progenitor numbers. Furthermore, RNAi dosing experiments indicate that progenitors closer to their normal target are relatively more sensitive to the effects of map3k1, implicating this factors in controlling the site of terminal differentiation. Eye phenotypes were also observed after inhibition of map2k4, map2k7, jnk, and p38, identifying a putative pathway through which map3k1 prevents differentiation. Together, these results suggest that map3k1 regulates a novel control point in the eye regeneration pathway which suppresses the terminal differentiation of progenitors during their migration to target destinations.
Conflict of interest statement
Competing Interest Statement: The authors declare that they have no competing interests.
Figures

Similar articles
-
Src acts with WNT/FGFRL signaling to pattern the planarian anteroposterior axis.Development. 2022 Apr 1;149(7):dev200125. doi: 10.1242/dev.200125. Epub 2022 Mar 30. Development. 2022. PMID: 35297964 Free PMC article.
-
Meis1 Controls the Differentiation of Eye Progenitor Cells and the Formation of Posterior Poles during Planarian Regeneration.Int J Mol Sci. 2023 Feb 9;24(4):3505. doi: 10.3390/ijms24043505. Int J Mol Sci. 2023. PMID: 36834910 Free PMC article.
-
Principles of regeneration revealed by the planarian eye.Curr Opin Cell Biol. 2021 Dec;73:19-25. doi: 10.1016/j.ceb.2021.05.001. Epub 2021 Jun 13. Curr Opin Cell Biol. 2021. PMID: 34134046 Free PMC article. Review.
-
Wnt/Notum spatial feedback inhibition controls neoblast differentiation to regulate reversible growth of the planarian brain.Development. 2015 Dec 15;142(24):4217-29. doi: 10.1242/dev.123612. Epub 2015 Nov 2. Development. 2015. PMID: 26525673 Free PMC article.
-
Genetic Control of MAP3K1 in Eye Development and Sex Differentiation.Cells. 2021 Dec 23;11(1):34. doi: 10.3390/cells11010034. Cells. 2021. PMID: 35011600 Free PMC article. Review.
References
-
- Reddien PW, Sánchez Alvarado A. Fundamentals of planarian regeneration. Annu Rev Cell Dev Biol. 2004;20:725–57. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
