This is a preprint.
Automated model-free analysis of cryo-EM volume ensembles with SIREn
- PMID: 39415986
- PMCID: PMC11482773
- DOI: 10.1101/2024.10.08.617123
Automated model-free analysis of cryo-EM volume ensembles with SIREn
Abstract
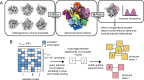
Cryogenic electron microscopy (cryo-EM) has the potential to capture snapshots of proteins in motion and generate hypotheses linking conformational states to biological function. This potential has been increasingly realized by the advent of machine learning models that allow 100s-1,000s of 3D density maps to be generated from a single dataset. How to identify distinct structural states within these volume ensembles and quantify their relative occupancies remain open questions. Here, we present an approach to inferring variable regions directly from a volume ensemble based on the statistical co-occupancy of voxels, as well as a 3D-convolutional neural network that predicts binarization thresholds for volumes in an unbiased and automated manner. We show that these tools recapitulate known heterogeneity in a variety of simulated and real cryo-EM datasets, and highlight how integrating these tools with existing data processing pipelines enables improved particle curation and the construction of quantitative conformational landscapes.
Conflict of interest statement
DECLARATION OF COMPETING INTERESTS The authors declare no competing interests.
Figures




Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
On the objectivity, reliability, and validity of deep learning enabled bioimage analyses.Elife. 2020 Oct 19;9:e59780. doi: 10.7554/eLife.59780. Elife. 2020. PMID: 33074102 Free PMC article.
-
Dynamic Field Theory of Executive Function: Identifying Early Neurocognitive Markers.Monogr Soc Res Child Dev. 2024 Dec;89(3):7-109. doi: 10.1111/mono.12478. Monogr Soc Res Child Dev. 2024. PMID: 39628288 Free PMC article.
-
Exploring conceptual and theoretical frameworks for nurse practitioner education: a scoping review protocol.JBI Database System Rev Implement Rep. 2015 Oct;13(10):146-55. doi: 10.11124/jbisrir-2015-2150. JBI Database System Rev Implement Rep. 2015. PMID: 26571290
-
Ethics of Procuring and Using Organs or Tissue from Infants and Newborns for Transplantation, Research, or Commercial Purposes: Protocol for a Bioethics Scoping Review.Wellcome Open Res. 2024 Dec 5;9:717. doi: 10.12688/wellcomeopenres.23235.1. eCollection 2024. Wellcome Open Res. 2024. PMID: 39839977 Free PMC article.
References
-
- Campello RJGB, Moulavi D, Zimek A, Sander J. 2015. Hierarchical Density Estimates for Data Clustering, Visualization, and Outlier Detection. ACM Transactions on Knowledge Discovery from Data (TKDD) 10.
-
- Cordasco G, Gargano L. 2010. Community detection via semi-synchronous label propagation algorithms. 2010 IEEE International Workshop on Business Applications of Social Network Analysis, BASNA 2010.
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
