Physiological, biochemical, and transcriptomic alterations in Castor (Ricinus communis L.) under polyethylene glycol-induced oxidative stress
- PMID: 39415088
- PMCID: PMC11484386
- DOI: 10.1186/s12870-024-05691-4
Physiological, biochemical, and transcriptomic alterations in Castor (Ricinus communis L.) under polyethylene glycol-induced oxidative stress
Abstract
Background: Castor is an important industrial raw material. Drought-induced oxidative stress leads to slow growth and decreased yields in castor. However, the mechanisms of drought-induced oxidative stress in castor remain unclear. Therefore, in this study, physiological, biochemical, and RNA-seq analyses were conducted on the roots of castor plants under PEG-6000 stress for 3 d and 7 d followed by 4 d of hydration.
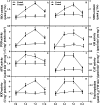
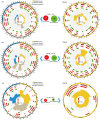
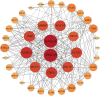
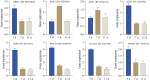
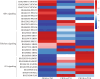
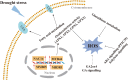
Results: The photosynthetic rate of castor leaves was inhibited under PEG-6000 stress for 3 and 7 d. Biochemical analysis of castor roots stressed for 3 d and 7 d, and rehydrated for 4 d revealed that the activities of APX and CAT were highest after only 3 d of stress, whereas the activities of POD, GR, and SOD peaked after 7 d of stress. RNA-seq analysis revealed 2926, 1507, and 111 differentially expressed genes (DEGs) in the roots of castor plants under PEG-6000 stress for 3 d and 7 d and after 4 d of rehydration, respectively. GO analysis of the DEGs indicated significant enrichment in antioxidant activity. Furthermore, KEGG enrichment analysis of the DEGs revealed significantly enriched metabolic pathways, including glutathione metabolism, fatty acid metabolism, and plant hormone signal transduction. WGCNA identified the core genes PP2C39 and GA2ox4 in the navajowhite1 module, which was upregulated under PEG-6000 stress. On the basis of these results, we propose a model for the response to drought-induced oxidative stress in castor.
Conclusions: This study provides valuable antioxidant gene resources, deepening our understanding of antioxidant regulation and paving the way for further molecular breeding of castor plants.
Keywords: Castor; PEG stress; RNA-seq.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


Similar articles
-
Genome-wide identification of core components of ABA signaling and transcriptome analysis reveals gene circuits involved in castor bean (Ricinus communis L.) response to drought.Gene. 2023 Oct 20;883:147668. doi: 10.1016/j.gene.2023.147668. Epub 2023 Jul 25. Gene. 2023. PMID: 37500024
-
Response of photosynthetic characteristics and antioxidant system in the leaves of safflower to NaCl and NaHCO3.Plant Cell Rep. 2024 May 20;43(6):146. doi: 10.1007/s00299-024-03234-7. Plant Cell Rep. 2024. PMID: 38764051
-
Castor (Ricinus communis L.) differential cell cycle and metabolism reactivation, germinability, and seedling performance under NaCl and PEG osmoticum: Stress tolerance related to genotype-preestablished superoxide dismutase activity.Plant Physiol Biochem. 2024 Feb;207:108372. doi: 10.1016/j.plaphy.2024.108372. Epub 2024 Jan 12. Plant Physiol Biochem. 2024. PMID: 38228015
-
Physiological and transcriptome analyses reveal tissue-specific responses of Leucaena plants to drought stress.Plant Physiol Biochem. 2024 Sep;214:108926. doi: 10.1016/j.plaphy.2024.108926. Epub 2024 Jul 9. Plant Physiol Biochem. 2024. PMID: 38996715
-
Piriformospora indica confers drought tolerance in Chinese cabbage leaves by stimulating antioxidant enzymes, the expression of drought-related genes and the plastid-localized CAS protein.J Plant Physiol. 2010 Aug 15;167(12):1009-17. doi: 10.1016/j.jplph.2010.02.013. Epub 2010 May 14. J Plant Physiol. 2010. PMID: 20471134
References
-
- Neeharika TSVR, Rani KNP, Rao KVSA, Kumar TP, Prasad RBN. Application of factorial design of experiments for the continuous hydrogenation of enriched castor oil methyl esters. Bull Chem Reaction Eng Catal. 2013;8(1978–2993):154–9.
-
- Nejeliski DM, Duarte LC. Waterproofing of bottle gourd (Lagenaria siceraria) with castor oil polyurethane resin. Materia-rio De Janeiro 2020, 25:2020.
-
- Yamamoto Y, Harada K, Kasuga S, Hosokawa M. Phospholipase A2-Mediated preparation of phosphatidylcholine containing ricinoleic acid and its anti-inflammatory effect on murine macrophage-like RAW264.7 cells. Biocatal Agric Biotechnol. 2019;19(1878–8181):101141–101141.
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

