Functional Analysis of Human GBA1 Missense Mutations in Drosophila: Insights into Gaucher Disease Pathogenesis and Phenotypic Consequences
- PMID: 39404383
- PMCID: PMC11475061
- DOI: 10.3390/cells13191619
Functional Analysis of Human GBA1 Missense Mutations in Drosophila: Insights into Gaucher Disease Pathogenesis and Phenotypic Consequences
Abstract
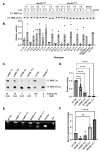
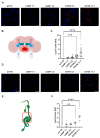
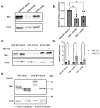
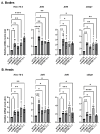
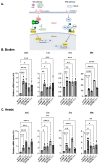
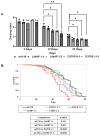
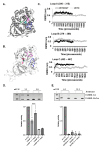
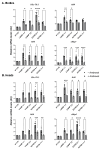
The human GBA1 gene encodes lysosomal acid β-glucocerebrosidase, whose activity is deficient in Gaucher disease (GD). In Drosophila, there are two GBA1 orthologs, Gba1a and Gba1b, and Gba1b is the bona fide GCase encoding gene. Several fly lines with different deletions in the Gba1b were studied in the past. However, since most GD-associated GBA1 mutations are point mutations, we created missense mutations homologous to the two most common GD mutations: the mild N370S mutation (D415S in Drosophila) and the severe L444P mutation (L494P in Drosophila), using the CRISPR-Cas9 technology. Flies homozygous for the D415S mutation (dubbed D370S hereafter) presented low GCase activity and substrate accumulation, which led to lysosomal defects, activation of the Unfolded Protein Response (UPR), inflammation/neuroinflammation, and neurodegeneration along with earlier death compared to control flies. Surprisingly, the L494P (called L444P hereafter) flies presented higher GCase activity with fewer lysosomal defects and milder disease in comparison to that presented by the D370S homozygous flies. Treatment with ambroxol had a limited effect on all homozygous fly lines tested. Overall, our results underscore the differences between the fly and human GCase enzymes, as evidenced by the distinct phenotypic outcomes of mutations in flies compared to those observed in human GD patients.
Keywords: Drosophila; GBA1; Gaucher disease; acid-β-glucocerebrosidase; ambroxol; glucosylceramide.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Gaucher Disease.2000 Jul 27 [updated 2023 Dec 7]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. 2000 Jul 27 [updated 2023 Dec 7]. In: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, editors. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. PMID: 20301446 Free Books & Documents. Review.
-
PGRN deficiency exacerbates, whereas a brain penetrant PGRN derivative protects, GBA1 mutation-associated pathologies and diseases.Proc Natl Acad Sci U S A. 2023 Jan 3;120(1):e2210442120. doi: 10.1073/pnas.2210442120. Epub 2022 Dec 27. Proc Natl Acad Sci U S A. 2023. PMID: 36574647 Free PMC article.
-
6-O-alkyl 4-methylumbelliferyl-β-D-glucosides as selective substrates for GBA1 in the discovery of glycosylated sterols.J Lipid Res. 2024 Nov;65(11):100670. doi: 10.1016/j.jlr.2024.100670. Epub 2024 Oct 10. J Lipid Res. 2024. PMID: 39395789 Free PMC article.
-
Erratum: Eyestalk Ablation to Increase Ovarian Maturation in Mud Crabs.J Vis Exp. 2023 May 26;(195). doi: 10.3791/6561. J Vis Exp. 2023. PMID: 37235796
-
Impact of residual disease as a prognostic factor for survival in women with advanced epithelial ovarian cancer after primary surgery.Cochrane Database Syst Rev. 2022 Sep 26;9(9):CD015048. doi: 10.1002/14651858.CD015048.pub2. Cochrane Database Syst Rev. 2022. PMID: 36161421 Free PMC article. Review.
References
-
- Dekker N., van Dussen L., Hollak C.E.M., Overkleeft H., Scheij S., Ghauharali K., van Breemen M.J., Ferraz M.J., Groener J.E.M., Maas M., et al. Elevated plasma glucosylsphingosine in Gaucher disease: Relation to phenotype, storage cell markers, and therapeutic response. Blood. 2011;118:e118–e127. doi: 10.1182/blood-2011-05-352971. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

