Intragenomic conflicts with plasmids and chromosomal mobile genetic elements drive the evolution of natural transformation within species
- PMID: 39401218
- PMCID: PMC11472951
- DOI: 10.1371/journal.pbio.3002814
Intragenomic conflicts with plasmids and chromosomal mobile genetic elements drive the evolution of natural transformation within species
Abstract
Natural transformation is the only mechanism of genetic exchange controlled by the recipient bacteria. We quantified its rates in 786 clinical strains of the human pathogens Legionella pneumophila (Lp) and 496 clinical and environmental strains of Acinetobacter baumannii (Ab). The analysis of transformation rates in the light of phylogeny revealed they evolve by a mixture of frequent small changes and a few large quick jumps across 6 orders of magnitude. In standard conditions close to half of the strains of Lp and a more than a third in Ab are below the detection limit and thus presumably non-transformable. Ab environmental strains tend to have higher transformation rates than the clinical ones. Transitions to non-transformability were frequent and usually recent, suggesting that they are deleterious and subsequently purged by natural selection. Accordingly, we find that transformation decreases genetic linkage in both species, which might accelerate adaptation. Intragenomic conflicts with chromosomal mobile genetic elements (MGEs) and plasmids could explain these transitions and a GWAS confirmed systematic negative associations between transformation and MGEs: plasmids and other conjugative elements in Lp, prophages in Ab, and transposable elements in both. In accordance with the hypothesis of modulation of transformation rates by genetic conflicts, transformable strains have fewer MGEs in both species and some MGEs inactivate genes implicated in the transformation with heterologous DNA (in Ab). Innate defense systems against MGEs are associated with lower transformation rates, especially restriction-modification systems. In contrast, CRISPR-Cas systems are associated with higher transformation rates suggesting that adaptive defense systems may facilitate cell protection from MGEs while preserving genetic exchanges by natural transformation. Ab and Lp have different lifestyles, gene repertoires, and population structure. Nevertheless, they exhibit similar trends in terms of variation of transformation rates and its determinants, suggesting that genetic conflicts could drive the evolution of natural transformation in many bacteria.
Copyright: © 2024 Mazzamurro et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
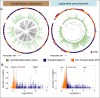
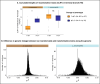



Similar articles
-
Diverse conjugative elements silence natural transformation in Legionella species.Proc Natl Acad Sci U S A. 2019 Sep 10;116(37):18613-18618. doi: 10.1073/pnas.1909374116. Epub 2019 Aug 27. Proc Natl Acad Sci U S A. 2019. PMID: 31455740 Free PMC article.
-
Horizontal DNA Transfer Mechanisms of Bacteria as Weapons of Intragenomic Conflict.PLoS Biol. 2016 Mar 2;14(3):e1002394. doi: 10.1371/journal.pbio.1002394. eCollection 2016 Mar. PLoS Biol. 2016. PMID: 26934590 Free PMC article.
-
CRISPR-Cas in mobile genetic elements: counter-defence and beyond.Nat Rev Microbiol. 2019 Aug;17(8):513-525. doi: 10.1038/s41579-019-0204-7. Nat Rev Microbiol. 2019. PMID: 31165781 Free PMC article.
-
Selfish, promiscuous and sometimes useful: how mobile genetic elements drive horizontal gene transfer in microbial populations.Philos Trans R Soc Lond B Biol Sci. 2022 Oct 10;377(1861):20210234. doi: 10.1098/rstb.2021.0234. Epub 2022 Aug 22. Philos Trans R Soc Lond B Biol Sci. 2022. PMID: 35989606 Free PMC article. Review.
-
Microbial evolution through horizontal gene transfer by mobile genetic elements.Microb Biotechnol. 2024 Jan;17(1):e14408. doi: 10.1111/1751-7915.14408. Epub 2024 Jan 16. Microb Biotechnol. 2024. PMID: 38226780 Free PMC article. Review.
References
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

