Thermal Denaturation of Fresh Frozen Tissue Enhances Mass Spectrometry Detection of Peptides
- PMID: 39392310
- PMCID: PMC11503521
- DOI: 10.1021/acs.analchem.4c03625
Thermal Denaturation of Fresh Frozen Tissue Enhances Mass Spectrometry Detection of Peptides
Abstract
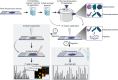
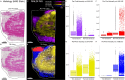
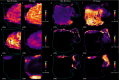
Thermal denaturation (TD), known as antigen retrieval, heats tissue samples in a buffered solution to expose protein epitopes. Thermal denaturation of formalin-fixed paraffin-embedded samples enhances on-tissue tryptic digestion, increasing peptide detection using matrix-assisted laser desorption ionization imaging mass spectrometry (MALDI IMS). We investigated the tissue-dependent effects of TD on peptide MALDI IMS and liquid chromatography-tandem mass spectrometry signal in unfixed, frozen human colon, ovary, and pancreas tissue. In a triplicate experiment using time-of-flight, orbitrap, and Fourier-transform ion cyclotron resonance mass spectrometry platforms, we found that TD had a tissue-dependent effect on peptide signal, resulting in a (22.5%) improvement in peptide detection from the colon, a (73.3%) improvement in ovary tissue, and a (96.6%) improvement in pancreas tissue. Biochemical analysis of identified peptides shows that TD facilitates identification of hydrophobic peptides.
Conflict of interest statement
The authors declare no competing financial interest.
Figures


Similar articles
-
In Situ Imaging of Tryptic Peptides by MALDI Imaging Mass Spectrometry Using Fresh-Frozen or Formalin-Fixed, Paraffin-Embedded Tissue.Curr Protoc Protein Sci. 2018 Nov;94(1):e65. doi: 10.1002/cpps.65. Epub 2018 Aug 16. Curr Protoc Protein Sci. 2018. PMID: 30114342 Free PMC article.
-
Citric acid antigen retrieval (CAAR) for tryptic peptide imaging directly on archived formalin-fixed paraffin-embedded tissue.J Proteome Res. 2010 Sep 3;9(9):4315-28. doi: 10.1021/pr9011766. J Proteome Res. 2010. PMID: 20597554
-
Recombinant " IMS TAG" proteins--a new method for validating bottom-up matrix-assisted laser desorption/ionisation ion mobility separation mass spectrometry imaging.Rapid Commun Mass Spectrom. 2013 Nov 15;27(21):2355-62. doi: 10.1002/rcm.6693. Rapid Commun Mass Spectrom. 2013. PMID: 24097391
-
Matrix-assisted laser desorption/ionization imaging mass spectrometry.Int J Mol Sci. 2010;11(12):5040-55. doi: 10.3390/ijms11125040. Epub 2010 Dec 7. Int J Mol Sci. 2010. PMID: 21614190 Free PMC article. Review.
-
MALDI imaging mass spectrometry and analysis of endogenous peptides.Expert Rev Proteomics. 2013 Aug;10(4):381-8. doi: 10.1586/14789450.2013.814939. Expert Rev Proteomics. 2013. PMID: 23992420 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

