Machine learning-guided reconstruction of cytoskeleton network from live-cell AFM images
- PMID: 39391724
- PMCID: PMC11465173
- DOI: 10.1016/j.isci.2024.110907
Machine learning-guided reconstruction of cytoskeleton network from live-cell AFM images
Abstract
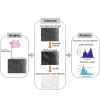
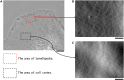
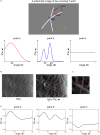
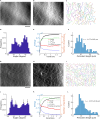
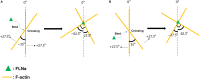
How actin filaments (F-actins) are dynamically reorganized in motile cells at the level of individual filaments is an open question. To find the answer, a high-speed atomic force microscopy (HS-AFM) system has been developed to live-imagine intracellular dynamics of the individual F-actins. However, noise and low resolution made it difficult to fully recognize individual F-actins in the HS-AFM images. To tackle this problem, we developed a new machine learning method that quantitatively recognizes individual F-actins. The method estimates F-actin orientation from the image while improving the resolution. We found that F-actins were oriented at ±35° toward the membrane in the lamellipodia, which is consistent with Arp2/3 complex-induced branching. Furthermore, in the cell cortex our results showed non-random orientation at four specific angles, suggesting a new mechanism for F-actin organization demonstrating the potential of our newly developed method to fundamentally improve our understanding of the structural dynamics of F-actin networks.
Keywords: biological sciences; computer science; engineering; physics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Leading edge maintenance in migrating cells is an emergent property of branched actin network growth.Elife. 2022 Mar 11;11:e74389. doi: 10.7554/eLife.74389. Elife. 2022. PMID: 35275060 Free PMC article.
-
In vivo dynamics of the cortical actin network revealed by fast-scanning atomic force microscopy.Microscopy (Oxf). 2017 Aug 1;66(4):272-282. doi: 10.1093/jmicro/dfx015. Microscopy (Oxf). 2017. PMID: 28531263
-
Probing in vivo dynamics of mitochondria and cortical actin networks using high-speed atomic force/fluorescence microscopy.Genes Cells. 2015 Feb;20(2):85-94. doi: 10.1111/gtc.12204. Epub 2014 Nov 30. Genes Cells. 2015. PMID: 25440894
-
Investigating the morphological dynamics of the plasma membrane by high-speed atomic force microscopy.J Cell Sci. 2021 Sep 1;134(17):jcs243584. doi: 10.1242/jcs.243584. Epub 2021 Sep 1. J Cell Sci. 2021. PMID: 34468000 Review.
-
The stabilization of Arp2/3 complex generated actin filaments.Biochem Soc Trans. 2024 Feb 28;52(1):343-352. doi: 10.1042/BST20230638. Biochem Soc Trans. 2024. PMID: 38288872 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

