Tissue-specific knockout in the Drosophila neuromuscular system reveals ESCRT's role in formation of synapse-derived extracellular vesicles
- PMID: 39388480
- PMCID: PMC11495600
- DOI: 10.1371/journal.pgen.1011438
Tissue-specific knockout in the Drosophila neuromuscular system reveals ESCRT's role in formation of synapse-derived extracellular vesicles
Abstract
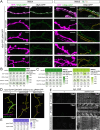
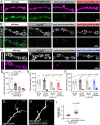
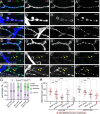
Tissue-specific gene knockout by CRISPR/Cas9 is a powerful approach for characterizing gene functions during development. However, this approach has not been successfully applied to most Drosophila tissues, including the Drosophila neuromuscular junction (NMJ). To expand tissue-specific CRISPR to this powerful model system, here we present a CRISPR-mediated tissue-restricted mutagenesis (CRISPR-TRiM) toolkit for knocking out genes in motoneurons, muscles, and glial cells. We validated the efficacy of CRISPR-TRiM by knocking out multiple genes in each tissue, demonstrated its orthogonal use with the Gal4/UAS binary expression system, and showed simultaneous knockout of multiple redundant genes. We used CRISPR-TRiM to discover an essential role for SNARE components in NMJ maintenance. Furthermore, we demonstrate that the canonical ESCRT pathway suppresses NMJ bouton growth by downregulating retrograde Gbb signaling. Lastly, we found that axon termini of motoneurons rely on ESCRT-mediated intra-axonal membrane trafficking to release extracellular vesicles at the NMJ. Thus, we have successfully developed an NMJ CRISPR mutagenesis approach which we used to reveal genes important for NMJ structural plasticity.
Copyright: © 2024 Chen et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Update of
-
Tissue-specific knockout in Drosophila neuromuscular system reveals ESCRT's role in formation of synapse-derived extracellular vesicles.bioRxiv [Preprint]. 2023 Sep 25:2023.09.25.559303. doi: 10.1101/2023.09.25.559303. bioRxiv. 2023. Update in: PLoS Genet. 2024 Oct 10;20(10):e1011438. doi: 10.1371/journal.pgen.1011438. PMID: 37808853 Free PMC article. Updated. Preprint.
Similar articles
-
Tissue-specific knockout in Drosophila neuromuscular system reveals ESCRT's role in formation of synapse-derived extracellular vesicles.bioRxiv [Preprint]. 2023 Sep 25:2023.09.25.559303. doi: 10.1101/2023.09.25.559303. bioRxiv. 2023. Update in: PLoS Genet. 2024 Oct 10;20(10):e1011438. doi: 10.1371/journal.pgen.1011438. PMID: 37808853 Free PMC article. Updated. Preprint.
-
The BMP homolog Gbb provides a retrograde signal that regulates synaptic growth at the Drosophila neuromuscular junction.Neuron. 2003 Jul 17;39(2):241-54. doi: 10.1016/s0896-6273(03)00426-4. Neuron. 2003. PMID: 12873382
-
Integration of a retrograde signal during synapse formation by glia-secreted TGF-β ligand.Curr Biol. 2012 Oct 9;22(19):1831-8. doi: 10.1016/j.cub.2012.07.063. Epub 2012 Sep 6. Curr Biol. 2012. PMID: 22959350 Free PMC article.
-
Development and plasticity of the Drosophila larval neuromuscular junction.Wiley Interdiscip Rev Dev Biol. 2013 Sep-Oct;2(5):647-70. doi: 10.1002/wdev.108. Epub 2013 Feb 5. Wiley Interdiscip Rev Dev Biol. 2013. PMID: 24014452 Free PMC article. Review.
-
Homeostatic plasticity at the Drosophila neuromuscular junction.Neuropharmacology. 2014 Mar;78:63-74. doi: 10.1016/j.neuropharm.2013.06.015. Epub 2013 Jun 24. Neuropharmacology. 2014. PMID: 23806804 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

