This is a preprint.
A one-step protocol to generate impermeable fluorescent HaloTag substrates for in situ live cell application and super-resolution imaging
- PMID: 39386703
- PMCID: PMC11463609
- DOI: 10.1101/2024.09.20.614087
A one-step protocol to generate impermeable fluorescent HaloTag substrates for in situ live cell application and super-resolution imaging
Abstract
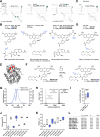
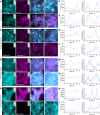
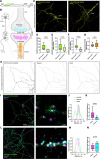
Communication between cells is largely orchestrated by proteins on the cell surface, which allow information transfer across the cell membrane. Super-resolution and single-molecule visualization of these proteins can be achieved by genetically grafting HTP (HaloTag Protein) into the protein of interest followed by brief incubation of cells with a dye-HTL (dye-linked HaloTag Ligand). This approach allows for use of cutting-edge fluorophores optimized for specific optical techniques or a cell-impermeable dye-HTL to selectively label surface proteins without labeling intracellular copies. However, these two goals often conflict, as many high-performing dyes exhibit membrane permeability. Traditional methods to eliminate cell permeability face synthetic bottlenecks and risk altering photophysical properties. Here we report that dye-HTL reagents can be made cell-impermeable by inserting a charged sulfonate directly into the HTL, leaving the dye moiety unperturbed. This simple, one-step method requires no purification and is compatible with both the original HTL and second-generation HTL.2, the latter offering accelerated labeling. We validate such compounds, termed dye-SHTL ('dye shuttle') conjugates, in live cells via widefield microscopy, demonstrating exclusive membrane staining of extracellular HTP fusion proteins. In transduced primary hippocampal neurons, we label mGluR2, a neuromodulatory G protein-coupled receptor (GPCR), with dyes optimized for stimulated emission by depletion (STED) super-resolution microscopy, allowing unprecedented accuracy in distinguishing surface and receptors from those in internal compartments of the presynaptic terminal, important in neural communication. This approach offers broad utility for surface-specific protein labelling.
Keywords: ATTO 647N; HaloTag; STED; fluorescence microscopy; impermeability; mGluR2.
Conflict of interest statement
COMPETING INTERESTS NL is a member of the scientific advisory board of Trace Neuroscience. MRT and BCS are on patent applications describing HTL.2. All other authors declare no competing interests.
Figures



Similar articles
-
Super-Resolution Fluorescence Imaging of Arabidopsis thaliana Transfer Cell Wall Ingrowths using Pseudo-Schiff Labelling Adapted for the Use of Different Dyes.Plant Cell Physiol. 2020 Oct 1;61(10):1775-1787. doi: 10.1093/pcp/pcaa102. Plant Cell Physiol. 2020. PMID: 32761075
-
Exchangeable HaloTag Ligands for Super-Resolution Fluorescence Microscopy.J Am Chem Soc. 2023 Feb 8;145(5):3075-3083. doi: 10.1021/jacs.2c11969. Epub 2023 Jan 30. J Am Chem Soc. 2023. PMID: 36716211 Free PMC article.
-
HaloTag technology for specific and covalent labeling of fusion proteins.Methods Mol Biol. 2015;1266:119-28. doi: 10.1007/978-1-4939-2272-7_8. Methods Mol Biol. 2015. PMID: 25560071
-
[Comparison and progress review of various super-resolution fluorescence imaging techniques].Se Pu. 2021 Oct;39(10):1055-1064. doi: 10.3724/SP.J.1123.2021.06015. Se Pu. 2021. PMID: 34505427 Free PMC article. Review. Chinese.
-
Fluorescent Probes for STED Optical Nanoscopy.Nanomaterials (Basel). 2021 Dec 22;12(1):21. doi: 10.3390/nano12010021. Nanomaterials (Basel). 2021. PMID: 35009972 Free PMC article. Review.
References
-
- Bosch P.J., Corrêa I.R., Sonntag M.H., Ibach J., Brunsveld L., Kanger J.S., and Subramaniam V. (2014). Evaluation of Fluorophores to Label SNAP-Tag Fused Proteins for Multicolor Single-Molecule Tracking Microscopy in Live Cells. Biophysical Journal 107, 803–814. 10.1016/j.bpj.2014.06.040. - DOI - PMC - PubMed
-
- Erdmann R.S., Baguley S.W., Richens J.H., Wissner R.F., Xi Z., Allgeyer E.S., Zhong S., Thompson A.D., Lowe N., Butler R., et al. (2019). Labeling Strategies Matter for Super-Resolution Microscopy: A Comparison between HaloTags and SNAP-tags. Cell Chemical Biology 26, 584–592.e6. 10.1016/j.chembiol.2019.01.003. - DOI - PMC - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
