Natural selection on apical membrane antigen 1 (AMA1) of an emerging zoonotic malaria parasite Plasmodium inui
- PMID: 39384839
- PMCID: PMC11464719
- DOI: 10.1038/s41598-024-74785-8
Natural selection on apical membrane antigen 1 (AMA1) of an emerging zoonotic malaria parasite Plasmodium inui
Abstract
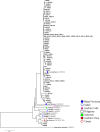
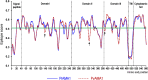
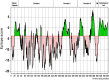
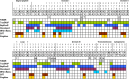
Apical membrane antigen 1 (AMA1) of malaria parasites plays an important role in host cell invasion. Antibodies to AMA1 can inhibit malaria merozoite invasion of erythrocytes while vaccine-induced specific cytotoxic T cell responses to this protein are associated with clinical protection. Polymorphisms in AMA1 of Plasmodium falciparum (PfAMA1) and P. vivax (PvAMA1) are of concern for vaccine development. To date, little is known about sequence diversity in ama1 of P. inui (Piama1), an emerging zoonotic malaria parasite. In this study, 80 complete Piama1 coding sequences were obtained from 57 macaques in Thailand that defined 60 haplotypes clustering in two phylogenetic lineages. In total, 74 nucleotide substitutions were identified and distributed unevenly across the gene. Blockwise analysis of the rates of synonymous (dS) and nonsynonymous (dN) nucleotide substitutions did not show a significant deviation from neutrality among Thai isolates. However, significantly negative Tajima's D values were detected in domain I and the loop region of domain II, implying purifying selection. Codon-based analysis of dN/dS has identified 12 and 14 codons under positive and negative selections, respectively. Meanwhile, 85 amino acid substitutions were identified among 80 Thai and 11 non-Thai PiAMA1 sequences. Of these, 48 substituted residues had a significant alteration in physicochemical properties, suggesting positive selection. More than half of these positively selected amino acids (32 of 48) corresponded to the predicted B-cell or T-cell epitopes, suggesting that selective pressure could be mediated by host immunity. Importantly, 14 amino acid substitutions were singletons and predicted to be deleterious that could be subject to ongoing purifying selection or elimination. Besides genetic drift and natural selection, intragenic recombination identified in domain II could generate sequence variation in Piama1. It is likely that malarial ama1 exhibits interspecies differences in evolutionary histories. Knowledge of the sequence diversity of the Piama1 locus further provides an evolutionary perspective of this important malaria vaccine candidate.
Keywords: Plasmodium inui; Apical membrane antigen 1; Malaria vaccine; Natural selection; Phenotypic plasticity; Zoonotic malaria.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Sequence diversity and natural selection at domain I of the apical membrane antigen 1 among Indian Plasmodium falciparum populations.Malar J. 2007 Nov 22;6:154. doi: 10.1186/1475-2875-6-154. Malar J. 2007. PMID: 18031585 Free PMC article.
-
Population genetics, sequence diversity and selection in the gene encoding the Plasmodium falciparum apical membrane antigen 1 in clinical isolates from the south-east of Iran.Infect Genet Evol. 2013 Jul;17:51-61. doi: 10.1016/j.meegid.2013.03.042. Epub 2013 Apr 2. Infect Genet Evol. 2013. PMID: 23557839
-
Structural diversity, natural selection and intragenic recombination in the Plasmodium vivax merozoite surface protein 9 locus in Thailand.Infect Genet Evol. 2020 Nov;85:104467. doi: 10.1016/j.meegid.2020.104467. Epub 2020 Jul 22. Infect Genet Evol. 2020. PMID: 32711079
-
Plasmodium vivax: sequence polymorphism and effect of natural selection at apical membrane antigen 1 (PvAMA1) among Indian population.Gene. 2008 Aug 1;419(1-2):35-42. doi: 10.1016/j.gene.2008.04.012. Epub 2008 Apr 28. Gene. 2008. PMID: 18547744
-
Apical membrane antigen 1: a malaria vaccine candidate in review.Trends Parasitol. 2008 Feb;24(2):74-84. doi: 10.1016/j.pt.2007.12.002. Epub 2008 Jan 15. Trends Parasitol. 2008. PMID: 18226584 Review.
References
-
- Cowman, A. F., Tonkin, C. J., Tham, W. H. & Duraisingh, M. T. The molecular basis of erythrocyte invasion by malaria parasites. Cell Host Microbe 22, 232–245 (2017). - PubMed
-
- Silvie, O. et al. A role for apical membrane antigen 1 during invasion of hepatocytes by Plasmodium falciparum sporozoites. J. Biol. Chem. 279, 9490–9496 (2004). - PubMed
-
- Collins, C. R. et al. Fine mapping of an epitope recognized by an invasion-inhibitory monoclonal antibody on the malaria vaccine candidate apical membrane antigen 1. J. Biol. Chem. 282, 7431–7441 (2007). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

