The middle domain of Hsp104 can ensure substrates are functional after processing
- PMID: 39361717
- PMCID: PMC11478891
- DOI: 10.1371/journal.pgen.1011424
The middle domain of Hsp104 can ensure substrates are functional after processing
Abstract
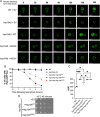
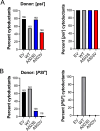
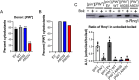
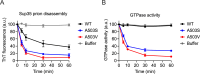
Molecular chaperones play a central role in protein disaggregation. However, the molecular determinants that regulate this process are poorly understood. Hsp104 is an AAA+ ATPase that disassembles stress granules and amyloids in yeast through collaboration with Hsp70 and Hsp40. In vitro studies show that Hsp104 processes different types of protein aggregates by partially translocating or threading polypeptides through the central pore of the hexamer. However, it is unclear how Hsp104 processing influences client protein function in vivo. The middle domain (MD) of Hsp104 regulates ATPase activity and interactions with Hsp70. Here, we tested how MD variants, Hsp104A503S and Hsp104A503V, process different protein aggregates. We establish that engineered MD variants fail to resolve stress granules but retain prion fragmentation activity required for prion propagation. Using the Sup35 prion protein, our in vitro and in vivo data indicate that the MD variants can disassemble Sup35 aggregates, but the disaggregated protein has reduced GTPase and translation termination activity. These results suggest that the middle domain can play a role in sensing certain substrates and plays an essential role in ensuring the processed protein is functional.
Copyright: © 2024 Buchholz et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
Exposed Hsp70-binding site impacts yeast Sup35 prion disaggregation and propagation.Proc Natl Acad Sci U S A. 2024 Dec 17;121(51):e2318162121. doi: 10.1073/pnas.2318162121. Epub 2024 Dec 10. Proc Natl Acad Sci U S A. 2024. PMID: 39656207 Free PMC article.
-
Molecular chaperone Hsp104 can promote yeast prion generation.Genetics. 2011 Jun;188(2):339-48. doi: 10.1534/genetics.111.127779. Epub 2011 Apr 5. Genetics. 2011. PMID: 21467567 Free PMC article.
-
Chaperone networks in protein disaggregation and prion propagation.J Struct Biol. 2012 Aug;179(2):152-60. doi: 10.1016/j.jsb.2012.05.002. Epub 2012 May 10. J Struct Biol. 2012. PMID: 22580344 Review.
-
Hsp70 targets Hsp100 chaperones to substrates for protein disaggregation and prion fragmentation.J Cell Biol. 2012 Aug 6;198(3):387-404. doi: 10.1083/jcb.201201074. J Cell Biol. 2012. PMID: 22869599 Free PMC article.
-
The life of [PSI].Curr Genet. 2018 Feb;64(1):1-8. doi: 10.1007/s00294-017-0714-7. Epub 2017 Jun 26. Curr Genet. 2018. PMID: 28653109 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

