N-terminal domain of androgen receptor is a major therapeutic barrier and potential pharmacological target for treating castration resistant prostate cancer: a comprehensive review
- PMID: 39359255
- PMCID: PMC11444995
- DOI: 10.3389/fphar.2024.1451957
N-terminal domain of androgen receptor is a major therapeutic barrier and potential pharmacological target for treating castration resistant prostate cancer: a comprehensive review
Abstract
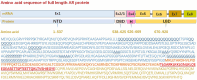
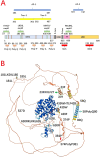
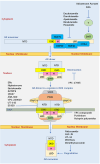
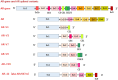
The incidence rate of prostate cancer (PCa) has risen by 3% per year from 2014 through 2019 in the United States. An estimated 34,700 people will die from PCa in 2023, corresponding to 95 deaths per day. Castration resistant prostate cancer (CRPC) is the leading cause of deaths among men with PCa. Androgen receptor (AR) plays a critical role in the development of CRPC. N-terminal domain (NTD) is the essential functional domain for AR transcriptional activation, in which modular activation function-1 (AF-1) is important for gene regulation and protein interactions. Over last 2 decades drug discovery against NTD has attracted interest for CRPC treatment. However, NTD is an intrinsically disordered domain without stable three-dimensional structure, which has so far hampered the development of drugs targeting this highly dynamic structure. Employing high throughput cell-based assays, small-molecule NTD inhibitors exhibit a variety of unexpected properties, ranging from specific binding to NTD, blocking AR transactivation, and suppressing oncogenic proliferation, which prompts its evaluation in clinical trials. Furthermore, molecular dynamics simulations reveal that compounds can induce the formation of collapsed helical states. Nevertheless, our knowledge of NTD structure has been limited to the primary sequence of amino acid chain and a few secondary structure motif, acting as a barrier for computational and pharmaceutical analysis to decipher dynamic conformation and drug-target interaction. In this review, we provide an overview on the sequence-structure-function relationships of NTD, including the polymorphism of mono-amino acid repeats, functional elements for transcription regulation, and modeled tertiary structure of NTD. Moreover, we summarize the activities and therapeutic potential of current NTD-targeting inhibitors and outline different experimental methods contributing to screening novel compounds. Finally, we discuss current directions for structure-based drug design and potential breakthroughs for exploring pharmacological motifs and pockets in NTD, which could contribute to the discovery of new NTD inhibitors.
Keywords: ARsplice variants; N-Terminal domain; androgen receptor; castration resistant prostate cancer; drug target.
Copyright © 2024 Chen and Lan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Sintokamide A Is a Novel Antagonist of Androgen Receptor That Uniquely Binds Activation Function-1 in Its Amino-terminal Domain.J Biol Chem. 2016 Oct 14;291(42):22231-22243. doi: 10.1074/jbc.M116.734475. Epub 2016 Aug 30. J Biol Chem. 2016. PMID: 27576691 Free PMC article.
-
Regression of castration-resistant prostate cancer by a novel compound QW07 targeting androgen receptor N-terminal domain.Cell Biol Toxicol. 2020 Oct;36(5):399-416. doi: 10.1007/s10565-020-09511-x. Epub 2020 Jan 30. Cell Biol Toxicol. 2020. PMID: 32002708
-
Androgen receptors in hormone-dependent and castration-resistant prostate cancer.Pharmacol Ther. 2013 Dec;140(3):223-38. doi: 10.1016/j.pharmthera.2013.07.003. Epub 2013 Jul 13. Pharmacol Ther. 2013. PMID: 23859952 Review.
-
Targeting the N-terminal domain of the androgen receptor: The effective approach in therapy of CRPC.Eur J Med Chem. 2023 Feb 5;247:115077. doi: 10.1016/j.ejmech.2022.115077. Epub 2022 Dec 30. Eur J Med Chem. 2023. PMID: 36587421 Review.
-
Targeting androgen receptor and the variants by an orally bioavailable Proteolysis Targeting Chimeras compound in castration resistant prostate cancer.EBioMedicine. 2023 Apr;90:104500. doi: 10.1016/j.ebiom.2023.104500. Epub 2023 Mar 7. EBioMedicine. 2023. PMID: 36893587 Free PMC article.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

