Development of novel canine phage display-derived neutralizing monoclonal antibody fragments against rabies virus from immunized dogs
- PMID: 39358469
- PMCID: PMC11447112
- DOI: 10.1038/s41598-024-73339-2
Development of novel canine phage display-derived neutralizing monoclonal antibody fragments against rabies virus from immunized dogs
Abstract
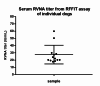
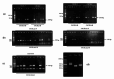
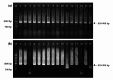
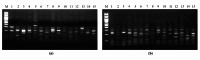
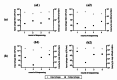
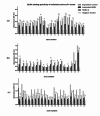
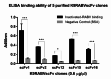
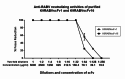
Animal rabies is a potentially fatal infectious disease in mammals, especially dogs. Currently, the number of rabies cases in pet dogs is increasing in several regions of Thailand. However, no passive postexposure prophylaxis (PEP) has been developed to combat rabies infection in animals. As monoclonal antibodies (MAbs) are promising biological therapies for postinfection, we developed a canine-neutralizing MAb against rabies virus (RABV) via the single-chain variable fragment (scFv) platform. Immunized phage-displaying scFv libraries were constructed from PBMCs via the pComb3XSS system. Diverse canine VHVLκ and VHVLλ libraries containing 2.4 × 108 and 1.3 × 106 clones, respectively, were constructed. Five unique clones that show binding affinity with the RABV glycoprotein were then selected, of which K9RABVscFv1 and K9RABVscFv16 showed rapid fluorescent foci inhibition test (RFFIT) neutralizing titers above the human protective level of 0.5 IU/ml. Finally, in silico docking predictions revealed that the residues on the CDRs of these neutralizing clones interact mainly with similar antigenic sites II and III on the RABV glycoprotein. These candidates may be used to develop complete anti-RABV MAbs as a novel PEP protocol in pet dogs and other animals.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
[Generation of Human ScFv Antibodies for Antigenic Site III of Rabies Virus Glycoprotein from Antibody-phage Libraries by Chain Shuffling].Bing Du Xue Bao. 2016 Jul;32(4):393-8. Bing Du Xue Bao. 2016. PMID: 29979522 Chinese.
-
Protective antibody response of Balb/c mice to Bali rabies virus isolate propagated in BHK-21 cells.J Vet Med Sci. 2018 Nov 1;80(10):1596-1603. doi: 10.1292/jvms.17-0385. Epub 2018 Sep 11. J Vet Med Sci. 2018. PMID: 30210066 Free PMC article.
-
Engineering of a recombinant trivalent single-chain variable fragment antibody directed against rabies virus glycoprotein G with improved neutralizing potency.Mol Immunol. 2014 Feb;57(2):66-73. doi: 10.1016/j.molimm.2013.08.009. Epub 2013 Oct 1. Mol Immunol. 2014. PMID: 24091293
-
A novel variable antibody fragment dimerized by leucine zippers with enhanced neutralizing potency against rabies virus G protein compared to its corresponding single-chain variable antibody fragment.Mol Immunol. 2015 Dec;68(2 Pt A):168-75. doi: 10.1016/j.molimm.2015.06.027. Epub 2015 Aug 29. Mol Immunol. 2015. PMID: 26325475
-
Negative effects of a disulfide bond mismatch in anti-rabies G protein single-chain antibody variable fragment FV57.Mol Immunol. 2014 Jun;59(2):136-41. doi: 10.1016/j.molimm.2014.01.006. Epub 2014 Mar 2. Mol Immunol. 2014. PMID: 24598312
References
-
- Niezgoda, M., Hanlon, C. A. & Rupprecht, C. E. Animal rabies. Rabies 163–218 (2003).
-
- Dietzschold, B., Tollis, M., Lafon, M., Wunner, W. H. & Koprowski, H. Mechanisms of Rabies virus neutralization by glycoprotein-specific monoclonal antibodies. Virology161, 29–36 (1987). - PubMed
-
- Flamand, V. et al. Vaccination with tumor-antigen-pulsed dendritic cells induces in vivo resistance to a B cell lymphoma. Adv. Exp. Med. Biol.329, 611–616. 10.1007/978-1-4615-2930-9_102 (1993). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

