Yeast Nat4 regulates DNA damage checkpoint signaling through its N-terminal acetyltransferase activity on histone H4
- PMID: 39356727
- PMCID: PMC11472955
- DOI: 10.1371/journal.pgen.1011433
Yeast Nat4 regulates DNA damage checkpoint signaling through its N-terminal acetyltransferase activity on histone H4
Abstract
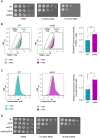
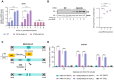
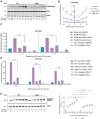
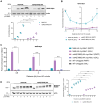
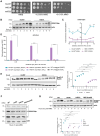
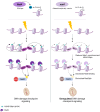
The DNA damage response (DDR) constitutes a vital cellular process that safeguards genome integrity. This biological process involves substantial alterations in chromatin structure, commonly orchestrated by epigenetic enzymes. Here, we show that the epigenetic modifier N-terminal acetyltransferase 4 (Nat4), known to acetylate the alpha-amino group of serine 1 on histones H4 and H2A, is implicated in the response to DNA damage in S. cerevisiae. Initially, we demonstrate that yeast cells lacking Nat4 have an increased sensitivity to DNA damage and accumulate more DNA breaks than wild-type cells. Accordingly, upon DNA damage, NAT4 gene expression is elevated, and the enzyme is specifically recruited at double-strand breaks. Delving deeper into its effects on the DNA damage signaling cascade, nat4-deleted cells exhibit lower levels of the damage-induced modification H2AS129ph (γH2A), accompanied by diminished binding of the checkpoint control protein Rad9 surrounding the double-strand break. Consistently, Mec1 kinase recruitment at double-strand breaks, critical for H2AS129ph deposition and Rad9 retention, is significantly impaired in nat4Δ cells. Consequently, Mec1-dependent phosphorylation of downstream effector kinase Rad53, indicative of DNA damage checkpoint activation, is reduced. Importantly, we found that the effects of Nat4 in regulating the checkpoint signaling cascade are mediated by its N-terminal acetyltransferase activity targeted specifically towards histone H4. Overall, this study points towards a novel functional link between histone N-terminal acetyltransferase Nat4 and the DDR, associating a new histone-modifying activity in the maintenance of genome integrity.
Copyright: © 2024 Constantinou et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Similar articles
-
DNA Damage-Induced Phosphorylation of Histone H2A at Serine 15 Is Linked to DNA End Resection.Mol Cell Biol. 2021 Nov 22;41(12):e0005621. doi: 10.1128/MCB.00056-21. Epub 2021 Sep 27. Mol Cell Biol. 2021. PMID: 34570618 Free PMC article.
-
Yeast G1 DNA damage checkpoint regulation by H2A phosphorylation is independent of chromatin remodeling.Proc Natl Acad Sci U S A. 2006 Sep 12;103(37):13771-6. doi: 10.1073/pnas.0511192103. Epub 2006 Aug 29. Proc Natl Acad Sci U S A. 2006. PMID: 16940359 Free PMC article.
-
Loss of Nat4 and its associated histone H4 N-terminal acetylation mediates calorie restriction-induced longevity.EMBO Rep. 2016 Dec;17(12):1829-1843. doi: 10.15252/embr.201642540. Epub 2016 Oct 31. EMBO Rep. 2016. PMID: 27799288 Free PMC article.
-
Interplays between ATM/Tel1 and ATR/Mec1 in sensing and signaling DNA double-strand breaks.DNA Repair (Amst). 2013 Oct;12(10):791-9. doi: 10.1016/j.dnarep.2013.07.009. Epub 2013 Aug 13. DNA Repair (Amst). 2013. PMID: 23953933 Review.
-
Yet another job for Dna2: Checkpoint activation.DNA Repair (Amst). 2015 Aug;32:17-23. doi: 10.1016/j.dnarep.2015.04.009. Epub 2015 May 1. DNA Repair (Amst). 2015. PMID: 25956863 Free PMC article. Review.
References
-
- Song O kyu, Wang X, Waterborg JH, Sternglanz R. An N α-Acetyltransferase Responsible for Acetylation of the N-terminal Residues of Histones H4 and H2A. J. Biol. Chem. 2003. Oct;278(40):38109–12. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

