Identification of common signature genes and pathways underlying the pathogenesis association between nonalcoholic fatty liver disease and heart failure
- PMID: 39351239
- PMCID: PMC11439677
- DOI: 10.3389/fimmu.2024.1424308
Identification of common signature genes and pathways underlying the pathogenesis association between nonalcoholic fatty liver disease and heart failure
Abstract
Background: Non-alcoholic fatty liver disease (NAFLD) and heart failure (HF) are related conditions with an increasing incidence. However, the mechanism underlying their association remains unclear. This study aimed to explore the shared pathogenic mechanisms and common biomarkers of NAFLD and HF through bioinformatics analyses and experimental validation.
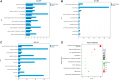
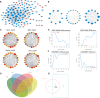
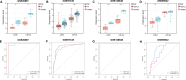
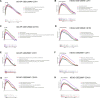
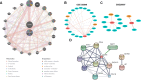
Methods: NAFLD and HF-related transcriptome data were extracted from the Gene Expression Omnibus (GEO) database (GSE126848 and GSE26887). Differential analysis was performed to identify common differentially expressed genes (co-DEGs) between NAFLD and HF. Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), and gene set enrichment analysis (GSEA) were conducted to explore the functions and regulatory pathways of co-DEGs. Protein-protein interaction (PPI) network and support vector machine-recursive feature elimination (SVM-RFE) methods were used to screen common key DEGs. The diagnostic value of common key DEGs was assessed by receiver operating characteristic (ROC) curve and validated with external datasets (GSE89632 and GSE57345). Finally, the expression of biomarkers was validated in mouse models.
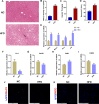
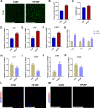
Results: A total of 161 co-DEGs were screened out in NAFLD and HF patients. GO, KEGG, and GSEA analyses indicated that these co-DEGs were mainly enriched in immune-related pathways. PPI network revealed 14 key DEGs, and SVM-RFE model eventually identified two genes (CD163 and CCR1) as common key DEGs for NAFLD and HF. Expression analysis revealed that the expression levels of CD163 and CCR1 were significantly down-regulated in HF and NAFLD patients. ROC curve analysis showed that CD163 and CCR1 had good diagnostic values for HF and NAFLD. Single-gene GSEA suggested that CD163 and CCR1 were mainly engaged in immune responses and inflammation. Experimental validation indicated unbalanced macrophage polarization in HF and NAFLD mouse models, and the expression of CD163 and CCR1 were significantly down-regulated.
Conclusion: This study identified M2 polarization impairment characterized by decreased expression of CD163 and CCR1 as a common pathogenic pathway in NAFLD and HF. The downregulation of CD163 and CCR1 may reflect key pathological changes in the development and progression of NAFLD and HF, suggesting their potential as diagnostic and therapeutic targets.
Keywords: bioinformatics; biomarker; heart failure; macrophage polarization; nonalcoholic fatty liver disease.
Copyright © 2024 Li, Lu and Chen.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Bioinformatics analysis reveals novel core genes associated with nonalcoholic fatty liver disease and nonalcoholic steatohepatitis.Gene. 2020 Jun 5;742:144549. doi: 10.1016/j.gene.2020.144549. Epub 2020 Mar 14. Gene. 2020. PMID: 32184169
-
Identification of candidate biomarkers and therapeutic agents for heart failure by bioinformatics analysis.BMC Cardiovasc Disord. 2021 Jul 4;21(1):329. doi: 10.1186/s12872-021-02146-8. BMC Cardiovasc Disord. 2021. PMID: 34218797 Free PMC article.
-
Identification of novel biomarkers, shared molecular signatures and immune cell infiltration in heart and kidney failure by transcriptomics.Front Immunol. 2024 Sep 16;15:1456083. doi: 10.3389/fimmu.2024.1456083. eCollection 2024. Front Immunol. 2024. PMID: 39351221 Free PMC article.
-
STAT4 and COL1A2 are potential diagnostic biomarkers and therapeutic targets for heart failure comorbided with depression.Brain Res Bull. 2022 Jun 15;184:68-75. doi: 10.1016/j.brainresbull.2022.03.014. Epub 2022 Mar 31. Brain Res Bull. 2022. PMID: 35367598 Review.
-
Effects of dietary curcumin on gene expression: An analysis of transcriptomic data in mice.Pathol Res Pract. 2024 Nov;263:155653. doi: 10.1016/j.prp.2024.155653. Epub 2024 Oct 11. Pathol Res Pract. 2024. PMID: 39426142 Review.
References
-
- Heidenreich PA, Bozkurt B, Aguilar D, Allen LA, Byun JJ, Colvin MM, et al. . Aha/Acc/Hfsa guideline for the management of heart failure: A report of the american college of cardiology/American heart association joint committee on clinical practice guidelines. Circulation. (2022) 145:e895–e1032. doi: 10.1161/CIR.0000000000001142 - DOI - PubMed
-
- Disease GBD, Injury I, Prevalence C. Global, regional, and national incidence, prevalence, and years lived with disability for 354 diseases and injuries for 195 countries and territories, 1990-2017: A systematic analysis for the global burden of disease study 2017. Lancet. (2018) 392:1789–858. doi: 10.1016/S0140-6736(18)32279-7 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

