QTL analysis to identify genes involved in the trade-off between silk protein synthesis and larva-pupa transition in silkworms
- PMID: 39350051
- PMCID: PMC11440889
- DOI: 10.1186/s12711-024-00937-z
QTL analysis to identify genes involved in the trade-off between silk protein synthesis and larva-pupa transition in silkworms
Abstract
Background: Insect-based food and feed are increasingly attracting attention. As a domesticated insect, the silkworm (Bombyx mori) has a highly nutritious pupa that can be easily raised in large quantities through large-scale farming, making it a highly promising source of food. The ratio of pupa to cocoon (RPC) refers to the proportion of the weight of the cocoon that is attributed to pupae, and is of significant value for edible utilization, as a higher RPC means a higher ratio of conversion of mulberry leaves to pupa. In silkworm production, there is a trade-off between RPC and cocoon shell ratiao(CSR), which refers the ratio of silk protein to the entire cocoon, during metamorphosis process. Understanding the genetic basis of this balance is crucial for breeding edible strains with a high RPC and further advancing its use as feed.
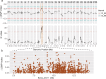
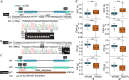
Results: Using QTL-seq, we identified a quantitative trait locus (QTL) for the balance between RPC and CSR that is located on chromosome 11 and covers a 9,773,115-bp region. This locus is an artificial selection hot spot that contains ten non-overlapping genomic regions under selection that were involved in the domestication and genetic breeding processes. These regions include 17 genes, nine of which are highly expressed in the silk gland, which is a vital component in the trade-off between RPC and CSR. These genes are annotate with function related with epigenetic modifications and the regulation of DNA replication et al. We identified one and two single nucleotide polymorphisms (SNPs) in the exons of teh KWMTBOMO06541 and KWMTBOMO06485 genes that result in amino acid changes in the protein domains. These SNPs have been strongly selected for during the domestication process. The KWMTBOMO06485 gene encodes the Bombyx mori (Bm) tRNA methyltransferase (BmDnmt2) and its knockout results in a significant change in the trade-off between CSR and RPC in both sexes.
Conclusions: Taken together, our results contribute to a better understanding of the genetic basis of RPC and CSR. The identified QTL and genes that affect RPC can be used for marker-assisted and genomic selection of silkworm strains with a high RPC. This will further enhance the production efficiency of silkworms and of closely-related insects for edible and feed purposes.
© 2024. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



Similar articles
-
Identification of Genes that Control Silk Yield by RNA Sequencing Analysis of Silkworm (Bombyx mori) Strains of Variable Silk Yield.Int J Mol Sci. 2018 Nov 22;19(12):3718. doi: 10.3390/ijms19123718. Int J Mol Sci. 2018. PMID: 30467288 Free PMC article.
-
Systemic disruption of the homeostasis of transfer RNA isopentenyltransferase causes growth and development abnormalities in Bombyx mori.Insect Mol Biol. 2019 Jun;28(3):380-391. doi: 10.1111/imb.12561. Epub 2019 Jan 24. Insect Mol Biol. 2019. PMID: 30548717
-
YorkieCA overexpression in the posterior silk gland improves silk yield in Bombyx mori.J Insect Physiol. 2017 Jul;100:93-99. doi: 10.1016/j.jinsphys.2017.06.001. Epub 2017 Jun 2. J Insect Physiol. 2017. PMID: 28583832
-
The beta-1, 4-N-acetylglucosaminidase 1 gene, selected by domestication and breeding, is involved in cocoon construction of Bombyx mori.PLoS Genet. 2020 Jul 15;16(7):e1008907. doi: 10.1371/journal.pgen.1008907. eCollection 2020 Jul. PLoS Genet. 2020. PMID: 32667927 Free PMC article.
-
Transgenic silkworms that weave recombinant proteins into silk cocoons.Biotechnol Lett. 2011 Apr;33(4):645-54. doi: 10.1007/s10529-010-0498-z. Epub 2010 Dec 24. Biotechnol Lett. 2011. PMID: 21184136 Review.
References
-
- Hawkey KJ, Lopez-Viso C, Brameld JM, Parr T, Salter AM. Insects: a potential source of protein and other nutrients for feed and food. Annu Rev Anim Biosci. 2021;9:333–54. - PubMed
-
- van Huis A. Potential of insects as food and feed in assuring food security. Annu Rev Entomol. 2013;58:563–83. - PubMed
-
- Wang YP, Liu J, Wu YM, Liu LE, Lv QJ, Wu YJ. Analysis of nutrition composition on silkworm pupa. J Zhengzhou Univ Med Sci. 2009;44:638–41.
-
- Köhler R, Kariuki L, Lambert C, Biesalski HK. Protein, amino acid and mineral composition of some edible insects from Thailand. J Asia Pac Entomol. 2019;22:372–8.
MeSH terms
Substances
Grants and funding
- 32330102/National Natural Science Foundation of China
- U20A2058/National Natural Science Foundation of China
- SWU-KQ22005/Fundamental Research Funds for the Central Universities
- cstc2021jcyj-cxtt0005/Natural Science Foundation of Chongqing, China
- cstc2021jcyj-bshX0014/Natural Science Foundation of Chongqing, China
LinkOut - more resources
Full Text Sources
Research Materials

