CHIMERA_NA: A Customizable Mutagenesis Tool for Structural Manipulations in Nucleic Acids and Their Complexes
- PMID: 39346815
- PMCID: PMC11425829
- DOI: 10.1021/acsomega.4c05954
CHIMERA_NA: A Customizable Mutagenesis Tool for Structural Manipulations in Nucleic Acids and Their Complexes
Abstract
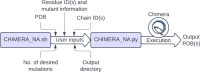
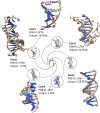
Studying the structure and dynamics of nucleic acids and their complexes is crucial for understanding fundamental biological processes and developing therapeutic interventions. However, the limited availability of experimentally characterized nucleic acid structures poses a challenge for exploring their properties comprehensively. To address this, we developed a customizable mutagenesis tool, CHIMERA_NA, to manipulate nucleic acid structures and their complexes. Utilizing the user-friendly CHIMERA_NA, researchers can perform mutations in nucleic acid structures, enabling the exploration of diverse structural configurations and dynamic behaviors. The tool offers the flexibility to generate all possible combinations of mutations or specific user-defined mutations based on research requirements. CHIMERA_NA leverages the capabilities of UCSF Chimera software, a widely used platform for molecular structure analysis, to facilitate the generation of mutations in nucleic acids. Our tool modifies the reference structure of nucleic acids or their complexes to generate initial coordinates of mutated structures/complexes within seconds for further computational exploration. This capability allows users to extend their investigations beyond structural repositories, enabling the study of DNA/RNA drug recognition, nucleic acid-protein interactions, and the intrinsic structural and dynamic properties of nucleic acids. By providing a user-friendly and customizable approach to nucleic acid mutagenesis, CHIMERA_NA contributes to advancing our understanding of nucleic acid biology and facilitating drug discovery efforts targeting nucleic acid-based mechanisms. CHIMERA_NA is freely available in the Supporting Information of this article.
© 2024 The Author. Published by American Chemical Society.
Conflict of interest statement
The author declares no competing financial interest.
Figures

Similar articles
-
Nucleic acid visualization with UCSF Chimera.Nucleic Acids Res. 2006 Feb 14;34(4):e29. doi: 10.1093/nar/gnj031. Nucleic Acids Res. 2006. PMID: 16478715 Free PMC article.
-
Tools for integrated sequence-structure analysis with UCSF Chimera.BMC Bioinformatics. 2006 Jul 12;7:339. doi: 10.1186/1471-2105-7-339. BMC Bioinformatics. 2006. PMID: 16836757 Free PMC article.
-
3D-NuS: A Web Server for Automated Modeling and Visualization of Non-Canonical 3-Dimensional Nucleic Acid Structures.J Mol Biol. 2017 Aug 4;429(16):2438-2448. doi: 10.1016/j.jmb.2017.06.013. Epub 2017 Jun 23. J Mol Biol. 2017. PMID: 28652006
-
Designer nucleic acids to probe and program the cell.Trends Cell Biol. 2012 Dec;22(12):624-33. doi: 10.1016/j.tcb.2012.10.001. Epub 2012 Nov 8. Trends Cell Biol. 2012. PMID: 23140833 Review.
-
Web tools support predicting protein-nucleic acid complexes stability with affinity changes.Wiley Interdiscip Rev RNA. 2023 Sep-Oct;14(5):e1781. doi: 10.1002/wrna.1781. Epub 2023 Jan 24. Wiley Interdiscip Rev RNA. 2023. PMID: 36693636 Review.
References
-
- Alberts B.; Johnson A.; Lewis J.; Raff M.; Roberts K.; Walter P., 2002. From DNA to RNA. In Mol. Biol. Cell. 4th ed. Garland Science.
-
- Elliott D.; Ladomery M., 2017. Molecular biology of RNA. Oxford University Press.
-
- Watson J. D.; Berry A., 2009. DNA: The secret of life. Knopf Doubleday Publishing Group.
-
- Neidle S.; Sanderson M., 2021. Principles of nucleic acid structure. Academic Press.
-
- Watson J. D.; Crick F. H., 1953, The structure of DNA. In Cold Spring Harbor symposia on quantitative biology (Vol. 18, pp 123–131). Cold Spring Harbor Laboratory Press. - PubMed
LinkOut - more resources
Full Text Sources
