This is a preprint.
Interphase chromosome conformation is specified by distinct folding programs inherited via mitotic chromosomes or through the cytoplasm
- PMID: 39345587
- PMCID: PMC11429855
- DOI: 10.1101/2024.09.16.613305
Interphase chromosome conformation is specified by distinct folding programs inherited via mitotic chromosomes or through the cytoplasm
Abstract
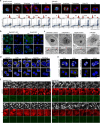
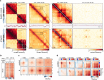
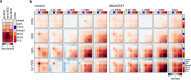
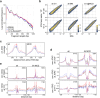
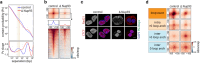
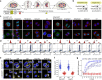
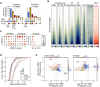
Identity-specific interphase chromosome conformation must be re-established each time a cell divides. To understand how interphase folding is inherited, we developed an experimental approach that physically segregates mediators of G1 folding that are intrinsic to mitotic chromosomes from cytoplasmic factors. Proteins essential for nuclear transport, RanGAP1 and Nup93, were degraded in pro-metaphase arrested DLD-1 cells to prevent the establishment of nucleo-cytoplasmic transport during mitotic exit and isolate the decondensing mitotic chromatin of G1 daughter cells from the cytoplasm. Using this approach, we discover a transient folding intermediate entirely driven by chromosome-intrinsic factors. In addition to conventional compartmental segregation, this chromosome-intrinsic folding program leads to prominent genome-scale microcompartmentalization of mitotically bookmarked and cell type-specific cis-regulatory elements. This microcompartment conformation is formed during telophase and subsequently modulated by a second folding program driven by factors inherited through the cytoplasm in G1. This nuclear import-dependent folding program includes cohesin and factors involved in transcription and RNA processing. The combined and inter-dependent action of chromosome-intrinsic and cytoplasmic inherited folding programs determines the interphase chromatin conformation as cells exit mitosis.
Conflict of interest statement
Conflicts of interest Job Dekker is a member of the scientific advisory board of Arima Genomics, San Diego, CA, USA and Omega Therapeutic, Cambridge, MA, USA.
Figures







Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
The effectiveness of school-based family asthma educational programs on the quality of life and number of asthma exacerbations of children aged five to 18 years diagnosed with asthma: a systematic review protocol.JBI Database System Rev Implement Rep. 2015 Oct;13(10):69-81. doi: 10.11124/jbisrir-2015-2335. JBI Database System Rev Implement Rep. 2015. PMID: 26571284
-
Can a Liquid Biopsy Detect Circulating Tumor DNA With Low-passage Whole-genome Sequencing in Patients With a Sarcoma? A Pilot Evaluation.Clin Orthop Relat Res. 2025 Jan 1;483(1):39-48. doi: 10.1097/CORR.0000000000003161. Epub 2024 Jun 21. Clin Orthop Relat Res. 2025. PMID: 38905450
-
Australia in 2030: what is our path to health for all?Med J Aust. 2021 May;214 Suppl 8:S5-S40. doi: 10.5694/mja2.51020. Med J Aust. 2021. PMID: 33934362
-
Interventions to reduce harm from continued tobacco use.Cochrane Database Syst Rev. 2016 Oct 13;10(10):CD005231. doi: 10.1002/14651858.CD005231.pub3. Cochrane Database Syst Rev. 2016. PMID: 27734465 Free PMC article. Review.
References
-
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T. 2001. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
