Human 8-oxoguanine glycosylase OGG1 binds nucleosome at the dsDNA ends and the super-helical locations
- PMID: 39341999
- PMCID: PMC11438860
- DOI: 10.1038/s42003-024-06919-7
Human 8-oxoguanine glycosylase OGG1 binds nucleosome at the dsDNA ends and the super-helical locations
Abstract
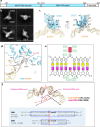
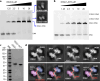
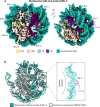
The human glycosylase OGG1 extrudes and excises the oxidized DNA base 8-oxoguanine (8-oxoG) to initiate base excision repair and plays important roles in many pathological conditions such as cancer, inflammation, and neurodegenerative diseases. Previous structural studies have used a truncated protein and short linear DNA, so it has been unclear how full-length OGG1 operates on longer DNA or on nucleosomes. Here we report cryo-EM structures of human OGG1 bound to a 35-bp long DNA containing an 8-oxoG within an unmethylated Cp-8-oxoG dinucleotide as well as to a nucleosome with an 8-oxoG at super-helical location (SHL)-5. The 8-oxoG in the linear DNA is flipped out by OGG1, consistent with previous crystallographic findings with a 15-bp DNA. OGG1 preferentially binds near dsDNA ends at the nucleosomal entry/exit sites. Such preference may underlie the enzyme's function in DNA double-strand break repair. Unexpectedly, we find that OGG1 bends the nucleosomal entry DNA, flips an undamaged guanine, and binds to internal nucleosomal DNA sites such as SHL-5 and SHL+6. We suggest that the DNA base search mechanism by OGG1 may be chromatin context-dependent and that OGG1 may partner with chromatin remodelers to excise 8-oxoG at the nucleosomal internal sites.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Lost in the Crowd: How Does Human 8-Oxoguanine DNA Glycosylase 1 (OGG1) Find 8-Oxoguanine in the Genome?Int J Mol Sci. 2020 Nov 7;21(21):8360. doi: 10.3390/ijms21218360. Int J Mol Sci. 2020. PMID: 33171795 Free PMC article. Review.
-
Base excision repair of 8-oxoG in dinucleosomes.Nucleic Acids Res. 2012 Jan;40(2):692-700. doi: 10.1093/nar/gkr761. Epub 2011 Sep 19. Nucleic Acids Res. 2012. PMID: 21930508 Free PMC article.
-
Human OGG1 activity in nucleosomes is facilitated by transient unwrapping of DNA and is influenced by the local histone environment.DNA Repair (Amst). 2017 Nov;59:1-8. doi: 10.1016/j.dnarep.2017.08.010. Epub 2017 Sep 1. DNA Repair (Amst). 2017. PMID: 28892740 Free PMC article.
-
Structural basis for the lack of opposite base specificity of Clostridium acetobutylicum 8-oxoguanine DNA glycosylase.DNA Repair (Amst). 2009 Nov 2;8(11):1283-9. doi: 10.1016/j.dnarep.2009.08.002. Epub 2009 Sep 10. DNA Repair (Amst). 2009. PMID: 19747886 Free PMC article.
-
Repair of 8-oxo-7,8-dihydroguanine in prokaryotic and eukaryotic cells: Properties and biological roles of the Fpg and OGG1 DNA N-glycosylases.Free Radic Biol Med. 2017 Jun;107:179-201. doi: 10.1016/j.freeradbiomed.2016.11.042. Epub 2016 Nov 27. Free Radic Biol Med. 2017. PMID: 27903453 Review.
References
-
- Friedberg, E. C., Walker, G. C., Siede, W. & Wood, R. D. DNA Repair and Mutagenesis (American Society for Microbiology Press, 2005).
-
- Friedberg, E. C. DNA damage and repair. Nature421, 436–440 (2003). - PubMed
-
- Burrows, C. J. Oxidative nucleobase modifications leading to strand scission. Chem. Rev.98, 1109–1151 (1998). - PubMed
MeSH terms
Substances
Grants and funding
- R01 ES011858/ES/NIEHS NIH HHS/United States
- R35 GM131754/GM/NIGMS NIH HHS/United States
- ES011858/U.S. Department of Health & Human Services | NIH | National Institute of Environmental Health Sciences (NIEHS)
- GM131754/U.S. Department of Health & Human Services | NIH | National Institute of General Medical Sciences (NIGMS)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

