The Autonomous Fusion Activity of Human Cytomegalovirus Glycoprotein B Is Regulated by Its Carboxy-Terminal Domain
- PMID: 39339958
- PMCID: PMC11437439
- DOI: 10.3390/v16091482
The Autonomous Fusion Activity of Human Cytomegalovirus Glycoprotein B Is Regulated by Its Carboxy-Terminal Domain
Abstract
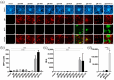
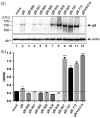
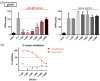
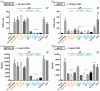
The human cytomegalovirus (HCMV) glycoprotein B (gB) is the viral fusogen required for entry into cells and for direct cell-to-cell spread of the virus. We have previously demonstrated that the exchange of the carboxy-terminal domain (CTD) of gB for the CTD of the structurally related fusion protein G of the vesicular stomatitis virus (VSV-G) resulted in an intrinsically fusion-active gB variant (gB/VSV-G). In this present study, we employed a dual split protein (DSP)-based cell fusion assay to further characterize the determinants of fusion activity in the CTD of gB. We generated a comprehensive library of gB CTD truncation mutants and identified two mutants, gB-787 and gB-807, which were fusion-competent and induced the formation of multinucleated cell syncytia in the absence of other HCMV proteins. Structural modeling coupled with site-directed mutagenesis revealed that gB fusion activity is primarily mediated by the CTD helix 2, and secondarily by the recruitment of cellular SH2/WW-domain-containing proteins. The fusion activity of gB-807 was inhibited by gB-specific monoclonal antibodies (MAbs) targeting the antigenic domains AD-1 to AD-5 within the ectodomain and not restricted to MAbs directed against AD-4 and AD-5 as observed for gB/VSV-G. This finding suggested a differential regulation of the fusion-active conformational state of both gB variants. Collectively, our findings underscore a pivotal role of the CTD in regulating the fusogenicity of HCMV gB, with important implications for understanding the conformations of gB that facilitate membrane fusion, including antigenic structures that could be targeted by antibodies to block this essential step in HCMV infection.
Keywords: blocking fusion; cell-cell fusion; gH/gL-independent fusion; glycoprotein B; herpesvirus; human cytomegalovirus; monoclonal antibodies.
Conflict of interest statement
The authors declare no commercial or financial conflicts of interest.
Figures


Similar articles
-
Human cytomegalovirus (HCMV) glycoprotein gB promotes virus entry in trans acting as the viral fusion protein rather than as a receptor-binding protein.mBio. 2013 Jun 4;4(3):e00332-13. doi: 10.1128/mBio.00332-13. mBio. 2013. PMID: 23736286 Free PMC article.
-
Monoclonal Antibodies to Different Components of the Human Cytomegalovirus (HCMV) Pentamer gH/gL/pUL128L and Trimer gH/gL/gO as well as Antibodies Elicited during Primary HCMV Infection Prevent Epithelial Cell Syncytium Formation.J Virol. 2016 Jun 24;90(14):6216-6223. doi: 10.1128/JVI.00121-16. Print 2016 Jul 15. J Virol. 2016. PMID: 27122579 Free PMC article.
-
In Vitro Viral Evolution Identifies a Critical Residue in the Alphaherpesvirus Fusion Glycoprotein B Ectodomain That Controls gH/gL-Independent Entry.mBio. 2021 May 4;12(3):e00557-21. doi: 10.1128/mBio.00557-21. mBio. 2021. PMID: 33947756 Free PMC article.
-
Pathogen at the Gates: Human Cytomegalovirus Entry and Cell Tropism.Viruses. 2018 Dec 11;10(12):704. doi: 10.3390/v10120704. Viruses. 2018. PMID: 30544948 Free PMC article. Review.
-
The COMPLEXity in herpesvirus entry.Curr Opin Virol. 2017 Jun;24:97-104. doi: 10.1016/j.coviro.2017.04.006. Epub 2017 May 21. Curr Opin Virol. 2017. PMID: 28538165 Free PMC article. Review.
References
-
- Teira P., Battiwalla M., Ramanathan M., Barrett A.J., Ahn K.W., Chen M., Green J.S., Saad A., Antin J.H., Savani B.N., et al. Early cytomegalovirus reactivation remains associated with increased transplant-related mortality in the current era: A CIBMTR analysis. Blood. 2016;127:2427–2438. doi: 10.1182/blood-2015-11-679639. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

