Exploring HIV-1 Maturation: A New Frontier in Antiviral Development
- PMID: 39339899
- PMCID: PMC11437483
- DOI: 10.3390/v16091423
Exploring HIV-1 Maturation: A New Frontier in Antiviral Development
Abstract
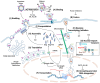
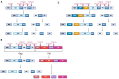
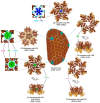
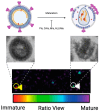
HIV-1 virion maturation is an essential step in the viral replication cycle to produce infectious virus particles. Gag and Gag-Pol polyproteins are assembled at the plasma membrane of the virus-producer cells and bud from it to the extracellular compartment. The newly released progeny virions are initially immature and noninfectious. However, once the Gag polyprotein is cleaved by the viral protease in progeny virions, the mature capsid proteins assemble to form the fullerene core. This core, harboring two copies of viral genomic RNA, transforms the virion morphology into infectious virus particles. This morphological transformation is referred to as maturation. Virion maturation influences the distribution of the Env glycoprotein on the virion surface and induces conformational changes necessary for the subsequent interaction with the CD4 receptor. Several host factors, including proteins like cyclophilin A, metabolites such as IP6, and lipid rafts containing sphingomyelins, have been demonstrated to have an influence on virion maturation. This review article delves into the processes of virus maturation and Env glycoprotein recruitment, with an emphasis on the role of host cell factors and environmental conditions. Additionally, we discuss microscopic technologies for assessing virion maturation and the development of current antivirals specifically targeting this critical step in viral replication, offering long-acting therapeutic options.
Keywords: Förster resonance energy transfer (FRET); Gag; Gag-Pol; allosteric integrase inhibitor (ALLINI); capsid; capsid inhibitor (CAI); human immunodeficiency virus type I (HIV-1); maturation; maturation inhibitor (MI); protease inhibitor (PI).
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Similar articles
-
Perturbing HIV-1 Ribosomal Frameshifting Frequency Reveals a cis Preference for Gag-Pol Incorporation into Assembling Virions.J Virol. 2022 Jan 12;96(1):e0134921. doi: 10.1128/JVI.01349-21. Epub 2021 Oct 13. J Virol. 2022. PMID: 34643428 Free PMC article.
-
Dominant Negative MA-CA Fusion Protein Is Incorporated into HIV-1 Cores and Inhibits Nuclear Entry of Viral Preintegration Complexes.J Virol. 2019 Oct 15;93(21):e01118-19. doi: 10.1128/JVI.01118-19. Print 2019 Nov 1. J Virol. 2019. PMID: 31413124 Free PMC article.
-
HIV-1 Maturation: Lessons Learned from Inhibitors.Viruses. 2020 Aug 26;12(9):940. doi: 10.3390/v12090940. Viruses. 2020. PMID: 32858867 Free PMC article. Review.
-
Subcellular Localization of HIV-1 gag-pol mRNAs Regulates Sites of Virion Assembly.J Virol. 2017 Feb 28;91(6):e02315-16. doi: 10.1128/JVI.02315-16. Print 2017 Mar 15. J Virol. 2017. PMID: 28053097 Free PMC article.
-
HIV-1 Capsid Inhibitors as Antiretroviral Agents.Curr HIV Res. 2016;14(3):270-82. doi: 10.2174/1570162x14999160224103555. Curr HIV Res. 2016. PMID: 26957201 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

