Exploring the structure and dynamics of soft and hard cuticle of Bombyx mori using solid-state NMR techniques
- PMID: 39333695
- PMCID: PMC11437033
- DOI: 10.1038/s41598-024-69984-2
Exploring the structure and dynamics of soft and hard cuticle of Bombyx mori using solid-state NMR techniques
Abstract
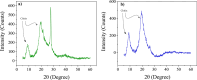
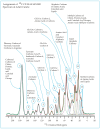
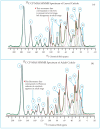
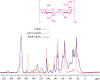
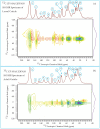
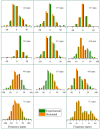
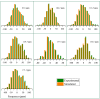
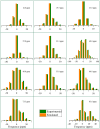
This study conducts a comprehensive analysis and comparison of Bombyx mori cuticles across different developmental stages, ranging from larval to adult, utilizing advanced solid-state NMR techniques. The primary objective is to elucidate the underlying reasons for the contrasting hardness of adult cuticles and softness of larval cuticles. Notably, PXRD analysis reveals a prominent broad peak at 19.34°, indicating the predominantly amorphous nature of both larval and adult cuticles. Analysis of 13C CP-MAS SSNMR spectra highlights an elevated proportion of phenoxy carbon in adult cuticles (6.77%) compared to larval cuticles (1.24%). Furthermore, a distinctive resonance line at 144 ppm is exclusively observed in adult cuticles, due to catechols, suggesting potential biochemical pathway variations during development. Significant variations in the primary components of 13C chemical shift anisotropy (CSA) tensors for aliphatic carbons of amino acids, catechols, and lipids between adult and larval cuticles indicate alterations in electronic environments. Additionally, the shorter spin-lattice relaxation time of carbon nuclei in larval cuticles compared to adult cuticles implies slower motional dynamics with enhanced degree of sclerotization in adults. By investigating the internal structure and dynamics of cuticles, this research not only contributes to biomimetic material development but also enhances our understanding of structural changes across different developmental stages of B. mori.
Keywords: Exoskeleton; Larval and adult cuticles of Bombyx mori; Solid-state NMR; XRD-measurements.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
A Blueprint of Microstructures and Stage-Specific Transcriptome Dynamics of Cuticle Formation in Bombyx mori.Int J Mol Sci. 2022 May 5;23(9):5155. doi: 10.3390/ijms23095155. Int J Mol Sci. 2022. PMID: 35563544 Free PMC article.
-
13C CP/MAS NMR study on structural heterogeneity in Bombyx mori silk fiber and their generation by stretching.Protein Sci. 2002 Nov;11(11):2706-13. doi: 10.1110/ps.0221702. Protein Sci. 2002. PMID: 12381852 Free PMC article.
-
Glycerin-Induced Conformational Changes in Bombyx mori Silk Fibroin Film Monitored by (13)C CP/MAS NMR and ¹H DQMAS NMR.Int J Mol Sci. 2016 Sep 9;17(9):1517. doi: 10.3390/ijms17091517. Int J Mol Sci. 2016. PMID: 27618034 Free PMC article.
-
Application of NMR microscopy to the morphological study of the silkworm, Bombyx mori, during its metamorphosis.Magn Reson Imaging. 1997;15(6):693-700. doi: 10.1016/s0730-725x(97)00006-4. Magn Reson Imaging. 1997. PMID: 9285809
-
Insect cuticular protein; gene expression, genomic structure, transcriptional regulation, speculated cuticular structure, clarified through the genomic analysis of Bombyx mori.Arch Insect Biochem Physiol. 2024 Aug;116(4):e22143. doi: 10.1002/arch.22143. Arch Insect Biochem Physiol. 2024. PMID: 39166352 Review.
References
-
- Moussian, B. Recent advances in understanding mechanisms of insect cuticle differentiation. Insect Biochem. Mol. Biol.40(5), 363–375 (2010). - PubMed
-
- Fraenkel, G. & Rudall, K. The structure of insect cuticles. Proc. Roy. Soc. Lond. Ser. B Biol. Sci.134(874), 111–143 (1947). - PubMed
-
- Resh, V. H. & Cardé, R. T. Encyclopedia of Insects (Academic Press, 2009).
-
- Kramer, K. J. & Hopkins, T. L. Tyrosine metabolism for insect cuticle tanning. Arch. Insect Biochem. Physiol.6(4), 279–301 (1987).
-
- Castejón, D., Rotllant, G., Ribes, E., Durfort, M. & Guerao, G. Structure of the stomach cuticle in adult and larvae of the spider crab Maja brachydactyla (Brachyura, Decapoda). J. Morphol.280(3), 370–380 (2019). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

