Disruption of the intestinal clock drives dysbiosis and impaired barrier function in colorectal cancer
- PMID: 39331712
- PMCID: PMC11430476
- DOI: 10.1126/sciadv.ado1458
Disruption of the intestinal clock drives dysbiosis and impaired barrier function in colorectal cancer
Abstract
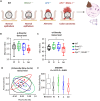
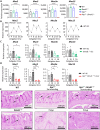
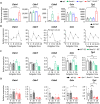
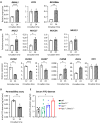
Diet is a robust entrainment cue that regulates diurnal rhythms of the gut microbiome. We and others have shown that disruption of the circadian clock drives the progression of colorectal cancer (CRC). While certain bacterial species have been suggested to play driver roles in CRC, it is unknown whether the intestinal clock impinges on the microbiome to accelerate CRC pathogenesis. To address this, genetic disruption of the circadian clock, in an Apc-driven mouse model of CRC, was used to define the impact on the gut microbiome. When clock disruption is combined with CRC, metagenomic sequencing identified dysregulation of many bacterial genera including Bacteroides, Helicobacter, and Megasphaera. We identify functional changes to microbial pathways including dysregulated nucleic acid, amino acid, and carbohydrate metabolism, as well as disruption of intestinal barrier function. Our findings suggest that clock disruption impinges on microbiota composition and intestinal permeability that may contribute to CRC pathogenesis.
Figures



Similar articles
-
High-Fat Diet Promotes Colorectal Tumorigenesis Through Modulating Gut Microbiota and Metabolites.Gastroenterology. 2022 Jan;162(1):135-149.e2. doi: 10.1053/j.gastro.2021.08.041. Epub 2021 Aug 27. Gastroenterology. 2022. PMID: 34461052
-
Disruption of the circadian clock drives Apc loss of heterozygosity to accelerate colorectal cancer.Sci Adv. 2022 Aug 12;8(32):eabo2389. doi: 10.1126/sciadv.abo2389. Epub 2022 Aug 10. Sci Adv. 2022. PMID: 35947664 Free PMC article.
-
The Circadian Clock Mutation Promotes Intestinal Dysbiosis.Alcohol Clin Exp Res. 2016 Feb;40(2):335-47. doi: 10.1111/acer.12943. Alcohol Clin Exp Res. 2016. PMID: 26842252 Free PMC article.
-
Metabolic Interaction Between Host and the Gut Microbiota During High-Fat Diet-Induced Colorectal Cancer.J Microbiol. 2024 Mar;62(3):153-165. doi: 10.1007/s12275-024-00123-2. Epub 2024 Apr 16. J Microbiol. 2024. PMID: 38625645 Review.
-
Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis.J Biomed Sci. 2018 Nov 9;25(1):79. doi: 10.1186/s12929-018-0483-8. J Biomed Sci. 2018. PMID: 30413188 Free PMC article. Review.
References
-
- Bass J., Lazar M. A., Circadian time signatures of fitness and disease. Science 354, 994–999 (2016). - PubMed
-
- Altman B. J., Hsieh A. L., Sengupta A., Krishnanaiah S. Y., Stine Z. E., Walton Z. E., Gouw A. M., Venkataraman A., Li B., Goraksha-Hicks P., Diskin S. J., Bellovin D. I., Simon M. C., Rathmell J. C., Lazar M. A., Maris J. M., Felsher D. W., Hogenesch J. B., Weljie A. M., Dang C. V., MYC disrupts the circadian clock and metabolism in cancer cells. Cell Metab. 22, 1009–1019 (2015). - PMC - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

