The Nucleolus and Its Interactions with Viral Proteins Required for Successful Infection
- PMID: 39329772
- PMCID: PMC11430610
- DOI: 10.3390/cells13181591
The Nucleolus and Its Interactions with Viral Proteins Required for Successful Infection
Abstract
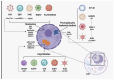
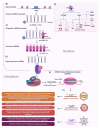
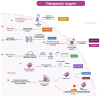
Nuclear bodies are structures in eukaryotic cells that lack a plasma membrane and are considered protein condensates, DNA, or RNA molecules. Known nuclear bodies include the nucleolus, Cajal bodies, and promyelocytic leukemia nuclear bodies. These bodies are involved in the concentration, exclusion, sequestration, assembly, modification, and recycling of specific components involved in the regulation of ribosome biogenesis, RNA transcription, and RNA processing. Additionally, nuclear bodies have been shown to participate in cellular processes such as the regulation of transcription of the cell cycle, mitosis, apoptosis, and the cellular stress response. The dynamics and functions of these bodies depend on the state of the cell. It is now known that both DNA and RNA viruses can direct their proteins to nuclear bodies, causing alterations in their composition, dynamics, and functions. Although many of these mechanisms are still under investigation, it is well known that the interaction between viral and nuclear body proteins is necessary for the success of the viral infection cycle. In this review, we concisely describe the interaction between viral and nuclear body proteins. Furthermore, we focus on the role of the nucleolus in RNA virus infections. Finally, we discuss the possible implications of the interaction of viral proteins on cellular transcription and the formation/degradation of non-coding RNAs.
Keywords: Cajal bodies; non-coding RNAs; nuclear bodies; nucleolus; promyelocytic leukemia nuclear bodies; ribosome biogenesis; transcription.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
Conventional and nonconventional roles of the nucleolus.Int Rev Cytol. 2002;219:199-266. doi: 10.1016/s0074-7696(02)19014-0. Int Rev Cytol. 2002. PMID: 12211630 Free PMC article. Review.
-
Nuclear bodies and compartments: functional roles and cellular signalling in health and disease.Cell Signal. 2004 Oct;16(10):1085-104. doi: 10.1016/j.cellsig.2004.03.020. Cell Signal. 2004. PMID: 15240004 Review.
-
Nucleolus: the fascinating nuclear body.Histochem Cell Biol. 2008 Jan;129(1):13-31. doi: 10.1007/s00418-007-0359-6. Epub 2007 Nov 29. Histochem Cell Biol. 2008. PMID: 18046571 Free PMC article. Review.
-
Viruses and the nucleolus: the fatal attraction.Biochim Biophys Acta. 2014 Jun;1842(6):840-7. doi: 10.1016/j.bbadis.2013.12.010. Epub 2013 Dec 27. Biochim Biophys Acta. 2014. PMID: 24378568 Free PMC article.
-
Manipulation of Cellular Processes via Nucleolus Hijaking in the Course of Viral Infection in Mammals.Cells. 2021 Jun 25;10(7):1597. doi: 10.3390/cells10071597. Cells. 2021. PMID: 34202380 Free PMC article. Review.
References
-
- Rawlinson S.M., Zhao T., Rozario A.M., Rootes C.L., McMillan P.J., Purcell A.W., Woon A., Marsh G.A., Lieu K.G., Wang L.-F., et al. Viral Regulation of Host Cell Biology by Hijacking of the Nucleolar DNA-Damage Response. Nat. Commun. 2018;9:3057. doi: 10.1038/s41467-018-05354-7. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

