Lactate-Induced HBEGF Shedding and EGFR Activation: Paving the Way to a New Anticancer Therapeutic Opportunity
- PMID: 39329717
- PMCID: PMC11430493
- DOI: 10.3390/cells13181533
Lactate-Induced HBEGF Shedding and EGFR Activation: Paving the Way to a New Anticancer Therapeutic Opportunity
Abstract
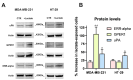
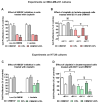
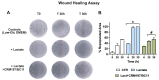
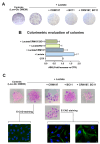
Cancer cells can release EGF-like peptides, acquiring the capacity of autocrine stimulation via EGFR-mediated signaling. One of these peptides (HBEGF) was found to be released from a membrane-bound precursor protein and is critically implicated in the proliferative potential of cancer cells. We observed that the increased lactate levels characterizing neoplastic tissues can induce the release of uPA, a protease promoting HBEGF shedding. This effect led to EGFR activation and increased ERK1/2 phosphorylation. Since EGFR-mediated signaling potentiates glycolytic metabolism, this phenomenon can induce a self-sustaining deleterious loop, favoring tumor growth. A well characterized HBEGF inhibitor is CRM197, a single-site variant of diphtheria toxin. We observed that, when administered individually, CRM197 did not trigger evident antineoplastic effects. However, its association with a uPA inhibitor caused dampening of EGFR-mediated signaling and apoptosis induction. Overall, our study highlights that the increased glycolytic metabolism and lactate production can foster the activated state of EGFR receptor and suggests that the inhibition of EGFR-mediated signaling can be attempted by means of CRM197 administered with an appropriate protease inhibitor. This attempt could help in overcoming the problem of the acquired resistance to the conventionally used EGFR inhibitors.
Keywords: EGFR; HBEGF; cancer cell metabolism; lactate.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures




Similar articles
-
Wound-induced HB-EGF ectodomain shedding and EGFR activation in corneal epithelial cells.Invest Ophthalmol Vis Sci. 2004 Mar;45(3):813-20. doi: 10.1167/iovs.03-0851. Invest Ophthalmol Vis Sci. 2004. PMID: 14985295 Free PMC article.
-
Antitumor effects of CRM197, a specific inhibitor of HB-EGF, in T-cell acute lymphoblastic leukemia.Anticancer Res. 2011 Jul;31(7):2483-8. Anticancer Res. 2011. PMID: 21873163
-
Enhancement of trophoblast differentiation and survival by low molecular weight heparin requires heparin-binding EGF-like growth factor.Hum Reprod. 2017 Jun 1;32(6):1218-1229. doi: 10.1093/humrep/dex069. Hum Reprod. 2017. PMID: 28402449 Free PMC article.
-
Human trophoblast survival at low oxygen concentrations requires metalloproteinase-mediated shedding of heparin-binding EGF-like growth factor.Development. 2006 Feb;133(4):751-9. doi: 10.1242/dev.02237. Epub 2006 Jan 11. Development. 2006. PMID: 16407398 Free PMC article.
-
Eicosanoids and HB-EGF/EGFR in cancer.Cancer Metastasis Rev. 2018 Sep;37(2-3):385-395. doi: 10.1007/s10555-018-9746-9. Cancer Metastasis Rev. 2018. PMID: 29936588 Review.
References
-
- Lorch J.H., Klessner J., Park J.K., Getsios S., Wu Y.L., Stack M.S., Green K.J. Epidermal growth factor receptor inhibition promotes desmosome assembly and strengthens intercellular adhesion in squamous cell carcinoma cells. J. Biol. Chem. 2004;279:37191–37200. doi: 10.1074/jbc.M405123200. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

