Fast and quantitative mitophagy assessment by flow cytometry using the mito-QC reporter
- PMID: 39324068
- PMCID: PMC11422238
- DOI: 10.3389/fcell.2024.1460061
Fast and quantitative mitophagy assessment by flow cytometry using the mito-QC reporter
Abstract
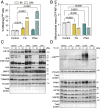
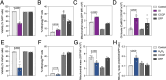
Mitochondrial quality control is finely tuned by mitophagy, the selective degradation of mitochondria through autophagy, and mitochondrial biogenesis. Removal of damaged mitochondria is essential to preserve cellular bioenergetics and prevent detrimental events such as sustained mitoROS production, pro-apoptotic cytochrome c release or mtDNA leakage. The array of tools available to study mitophagy is very limited but in constant development. Almost a decade ago, we developed a method to assess mitophagy flux using MitoTracker Deep Red in combination with lysosomal inhibitors. Now, using the novel tandem-fluorescence reporter mito-QC (mCherry-GFP-FIS1101-152) that allows to differentiate between healthy mitochondria (mCherry+GFP+) and mitolysosomes (mCherry+GFP-), we have developed a robust and quantitative method to assess mitophagy by flow cytometry. This approach has been validated in ARPE-19 cells using PINK1/Parkin-dependent (CCCP) and PINK1/Parkin-independent (DFP) positive controls and complementary techniques. Furthermore, we show that the mito-QC reporter can be multiplexed, especially if using spectral flow cytometry, to simultaneously study other cellular parameters such as viability or ROS production. Using this technique, we evaluated and characterized two prospective mitophagy inducers and further dissected their mechanism of action. Finally, using mito-QC reporter mice, we developed a protocol to measure mitophagy levels in the retina ex vivo. This novel methodology will propel mitophagy research forward and accelerate the discovery of novel mitophagy modulators.
Keywords: FACS; Fisetin; SI; autophagy; mitochondria; phenanthroline; retina.
Copyright © 2024 Jiménez-Loygorri, Jiménez-García, Viedma-Poyatos and Boya.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Mt-Keima detects PINK1-PRKN mitophagy in vivo with greater sensitivity than mito-QC.Autophagy. 2021 Nov;17(11):3753-3762. doi: 10.1080/15548627.2021.1896924. Epub 2021 Mar 8. Autophagy. 2021. PMID: 33685343 Free PMC article.
-
BNIP3L-mediated mitophagy is required for mitochondrial remodeling during the differentiation of optic nerve oligodendrocytes.Autophagy. 2021 Oct;17(10):3140-3159. doi: 10.1080/15548627.2020.1871204. Epub 2021 Jan 19. Autophagy. 2021. PMID: 33404293 Free PMC article.
-
Semi-automated quantitation of mitophagy in cells and tissues.Mech Ageing Dev. 2020 Jan;185:111196. doi: 10.1016/j.mad.2019.111196. Epub 2019 Dec 13. Mech Ageing Dev. 2020. PMID: 31843465 Free PMC article.
-
New methods for monitoring mitochondrial biogenesis and mitophagy in vitro and in vivo.Exp Biol Med (Maywood). 2017 Apr;242(8):781-787. doi: 10.1177/1535370216688802. Epub 2017 Jan 1. Exp Biol Med (Maywood). 2017. PMID: 28093935 Free PMC article. Review.
-
Hallmarks and Molecular Tools for the Study of Mitophagy in Parkinson's Disease.Cells. 2022 Jul 2;11(13):2097. doi: 10.3390/cells11132097. Cells. 2022. PMID: 35805181 Free PMC article. Review.
References
-
- Chan C. M., Huang D. Y., Sekar P., Hsu S. H., Lin W. W. (2019). Reactive oxygen species-dependent mitochondrial dynamics and autophagy confer protective effects in retinal pigment epithelial cells against sodium iodate-induced cell death. J. Biomed. Sci. 26 (1), 40. 10.1186/s12929-019-0531-z - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

