Comparative analysis of two independent Myh6-Cre transgenic mouse lines
- PMID: 39323506
- PMCID: PMC11423776
- DOI: 10.1016/j.jmccpl.2024.100081
Comparative analysis of two independent Myh6-Cre transgenic mouse lines
Abstract
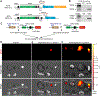
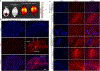
We have previously shown that the Myh6 promoter drives Cre expression in a subset of male germ line cells in three independent Myh6-Cre mouse lines, including two transgenic lines and one knock-in allele. In this study, we further compared the tissue-specificity of the two Myh6-Cre transgenic mouse lines, MDS Myh6-Cre and AUTR Myh6-Cre, through examining the expression of tdTomato (tdTom) red fluorescence protein in multiple internal organs, including the heart, brain, liver, lung, pancreas and brown adipose tissue. Our results show that MDS Myh6-Cre mainly activates tdTom reporter in the heart, whereas AUTR Myh6-Cre activates tdTom expression significantly in the heart, and in the cells of liver, pancreas and brain. In the heart, similar to MDS Myh6-Cre, AUTR Myh6-Cre activates tdTom in most cardiomyocytes. In the other organs, AUTR Myh6-Cre not only mosaically activates tdTom in some parenchymal cells, such as hepatocytes in the liver and neurons in the brain, but also turns on tdTom in some interstitial cells of unknown identity.
Keywords: Cre-loxP system; Genomic position effect; Myh6-Cre; Transgenic mouse line; tdTomato reporter.
Figures
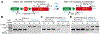



Similar articles
-
Myh6 promoter-driven Cre recombinase excises floxed DNA fragments in a subset of male germline cells.J Mol Cell Cardiol. 2023 Feb;175:62-66. doi: 10.1016/j.yjmcc.2022.12.005. Epub 2022 Dec 28. J Mol Cell Cardiol. 2023. PMID: 36584478 Free PMC article.
-
Generation and characterization of a Myh6-driven Cre knockin mouse line.Transgenic Res. 2021 Dec;30(6):821-835. doi: 10.1007/s11248-021-00285-4. Epub 2021 Sep 20. Transgenic Res. 2021. PMID: 34542814 Free PMC article.
-
A Murine Myh6MerCreMer Knock-In Allele Specifically Mediates Temporal Genetic Deletion in Cardiomyocytes after Tamoxifen Induction.PLoS One. 2015 Jul 23;10(7):e0133472. doi: 10.1371/journal.pone.0133472. eCollection 2015. PLoS One. 2015. PMID: 26204265 Free PMC article.
-
Establishment of Neh2-Cre:tdTomato reporter mouse for monitoring the exposure history to electrophilic stress.Free Radic Biol Med. 2022 Nov 20;193(Pt 2):610-619. doi: 10.1016/j.freeradbiomed.2022.11.004. Epub 2022 Nov 9. Free Radic Biol Med. 2022. PMID: 36368569 Review.
-
Conditional gene expression in the mouse inner ear using Cre-loxP.J Assoc Res Otolaryngol. 2012 Jun;13(3):295-322. doi: 10.1007/s10162-012-0324-5. Epub 2012 Apr 24. J Assoc Res Otolaryngol. 2012. PMID: 22526732 Free PMC article. Review.
References
-
- Ray MK, Fagan SP, Brunicardi FC. The Cre–loxP system: A versatile tool for targeting genes in a cell-and stage-specific manner. Cell Transplant 2000;9:805–15. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
