Fibroblast-derived extracellular vesicles contain SFRP1 and mediate pulmonary fibrosis
- PMID: 39315549
- PMCID: PMC11457858
- DOI: 10.1172/jci.insight.168889
Fibroblast-derived extracellular vesicles contain SFRP1 and mediate pulmonary fibrosis
Abstract
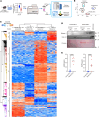
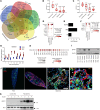
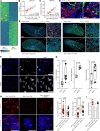
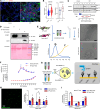
Idiopathic pulmonary fibrosis (IPF) is a lethal chronic lung disease characterized by aberrant intercellular communication, extracellular matrix deposition, and destruction of functional lung tissue. While extracellular vesicles (EVs) accumulate in the IPF lung, their cargo and biological effects remain unclear. We interrogated the proteome of EV and non-EV fractions during pulmonary fibrosis and characterized their contribution to fibrosis. EVs accumulated 14 days after bleomycin challenge, correlating with decreased lung function and initiated fibrogenesis in healthy precision-cut lung slices. Label-free proteomics of bronchoalveolar lavage fluid EVs (BALF-EVs) collected from mice challenged with bleomycin or control identified 107 proteins enriched in fibrotic vesicles. Multiomic analysis revealed fibroblasts as a major cellular source of BALF-EV cargo, which was enriched in secreted frizzled related protein 1 (SFRP1). Sfrp1 deficiency inhibited the activity of fibroblast-derived EVs to potentiate lung fibrosis in vivo. SFRP1 led to increased transitional cell markers, such as keratin 8, and WNT/β-catenin signaling in primary alveolar type 2 cells. SFRP1 was expressed within the IPF lung and localized at the surface of EVs from patient-derived fibroblasts and BALF. Our work reveals altered EV protein cargo in fibrotic EVs promoting fibrogenesis and identifies fibroblast-derived vesicular SFRP1 as a fibrotic mediator and potential therapeutic target for IPF.
Keywords: Cell biology; Fibrosis; Molecular biology; Pulmonology.
Figures


Similar articles
-
Increased Extracellular Vesicles Mediate WNT5A Signaling in Idiopathic Pulmonary Fibrosis.Am J Respir Crit Care Med. 2018 Dec 15;198(12):1527-1538. doi: 10.1164/rccm.201708-1580OC. Am J Respir Crit Care Med. 2018. PMID: 30044642
-
Human bronchial epithelial cell-derived extracellular vesicle therapy for pulmonary fibrosis via inhibition of TGF-β-WNT crosstalk.J Extracell Vesicles. 2021 Aug;10(10):e12124. doi: 10.1002/jev2.12124. Epub 2021 Aug 2. J Extracell Vesicles. 2021. PMID: 34377373 Free PMC article.
-
Mesenchymal stem cell-derived extracellular vesicles suppress the fibroblast proliferation by downregulating FZD6 expression in fibroblasts via micrRNA-29b-3p in idiopathic pulmonary fibrosis.J Cell Physiol. 2020 Nov;235(11):8613-8625. doi: 10.1002/jcp.29706. Epub 2020 Jun 17. J Cell Physiol. 2020. PMID: 32557673
-
The Role of Extracellular Vesicles in Idiopathic Pulmonary Fibrosis Progression: An Approach on Their Therapeutics Potential.Cells. 2022 Feb 11;11(4):630. doi: 10.3390/cells11040630. Cells. 2022. PMID: 35203281 Free PMC article. Review.
-
Extracellular Vesicles from Airway Secretions: New Insights in Lung Diseases.Int J Mol Sci. 2021 Jan 8;22(2):583. doi: 10.3390/ijms22020583. Int J Mol Sci. 2021. PMID: 33430153 Free PMC article. Review.
Cited by
-
Harmonising cellular conversations: decoding the vital roles of extracellular vesicles in respiratory system intercellular communications.Eur Respir Rev. 2024 Nov 13;33(174):230272. doi: 10.1183/16000617.0272-2023. Print 2024 Oct. Eur Respir Rev. 2024. PMID: 39537245 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

