This is a preprint.
Trehalose catalytic shift is an intrinsic factor in Mycobacterium tuberculosis that enhances phenotypic heterogeneity and multidrug resistance
- PMID: 39315249
- PMCID: PMC11419184
- DOI: 10.21203/rs.3.rs-4999164/v1
Trehalose catalytic shift is an intrinsic factor in Mycobacterium tuberculosis that enhances phenotypic heterogeneity and multidrug resistance
Abstract
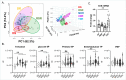
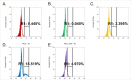
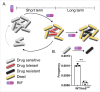
Drug-resistance (DR) in many bacterial pathogens often arises from the repetitive formation of drug-tolerant bacilli, known as persisters. However, it is unclear whether Mycobacterium tuberculosis (Mtb), the bacterium that causes tuberculosis (TB), undergoes a similar phenotypic transition. Recent metabolomics studies have identified that a change in trehalose metabolism is necessary for Mtb to develop persisters and plays a crucial role in metabolic networks of DR-TB strains. The present study used Mtb mutants lacking the trehalose catalytic shift and showed that the mutants exhibited a significantly lower frequency of the emergence of DR mutants compared to wildtype, due to reduced persister formation. The trehalose catalytic shift enables Mtb persisters to survive under bactericidal antibiotics by increasing metabolic heterogeneity and drug tolerance, ultimately leading to development of DR. Intriguingly, rifampicin (RIF)-resistant bacilli exhibit cross-resistance to a second antibiotic, due to a high trehalose catalytic shift activity. This phenomenon explains how the development of multidrug resistance (MDR) is facilitated by the acquisition of RIF resistance. In this context, the heightened risk of MDR-TB in the lineage 4 HN878 W-Beijing strain can be attributed to its greater trehalose catalytic shift. Genetic and pharmacological inactivation of the trehalose catalytic shift significantly reduced persister formation, subsequently decreasing the incidence of MDR-TB in HN878 W-Beijing strain. Collectively, the trehalose catalytic shift serves as an intrinsic factor of Mtb responsible for persister formation, cross-resistance to multiple antibiotics, and the emergence of MDR-TB. This study aids in the discovery of new TB therapeutics by targeting the trehalose catalytic shift of Mtb.
Figures




Similar articles
-
Central carbon metabolism remodeling as a mechanism to develop drug tolerance and drug resistance in Mycobacterium tuberculosis.Front Cell Infect Microbiol. 2022 Aug 22;12:958240. doi: 10.3389/fcimb.2022.958240. eCollection 2022. Front Cell Infect Microbiol. 2022. PMID: 36072228 Free PMC article. Review.
-
Transient drug-tolerance and permanent drug-resistance rely on the trehalose-catalytic shift in Mycobacterium tuberculosis.Nat Commun. 2019 Jul 2;10(1):2928. doi: 10.1038/s41467-019-10975-7. Nat Commun. 2019. PMID: 31266959 Free PMC article.
-
Targeting Mycobacterium tuberculosis Persistence through Inhibition of the Trehalose Catalytic Shift.ACS Infect Dis. 2024 Apr 12;10(4):1391-1404. doi: 10.1021/acsinfecdis.4c00138. Epub 2024 Mar 14. ACS Infect Dis. 2024. PMID: 38485491 Free PMC article.
-
Use of GeneXpert Mycobacterium tuberculosis/rifampicin for rapid detection of rifampicin resistant Mycobacterium tuberculosis strains of clinically suspected multi-drug resistance tuberculosis cases.Ann Transl Med. 2016 May;4(9):168. doi: 10.21037/atm.2016.05.09. Ann Transl Med. 2016. PMID: 27275481 Free PMC article.
-
Xpert® MTB/RIF assay for pulmonary tuberculosis and rifampicin resistance in adults.Cochrane Database Syst Rev. 2014 Jan 21;2014(1):CD009593. doi: 10.1002/14651858.CD009593.pub3. Cochrane Database Syst Rev. 2014. Update in: Cochrane Database Syst Rev. 2019 Jun 07;6:CD009593. doi: 10.1002/14651858.CD009593.pub4. PMID: 24448973 Free PMC article. Updated. Review.
References
-
- WHO. Global tuberculosis report (2022).
-
- Nathan C. Taming tuberculosis: a challenge for science and society. Cell Host Microbe 5, 220–224 (2009). - PubMed
-
- Nathan C. Drug-resistant tuberculosis: a new shot on goal. Nat Med 20, 121–123 (2014). - PubMed
-
- Gold B., Warrier T. & Nathan C. A multi-stress model for high throughput screening against non-replicating Mycobacterium tuberculosis. Methods Mol Biol 1285, 293–315 (2015). - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources

