This is a preprint.
Light-triggered protease-mediated release of actin-bound cargo from synthetic cells
- PMID: 39314483
- PMCID: PMC11419145
- DOI: 10.1101/2024.09.15.613133
Light-triggered protease-mediated release of actin-bound cargo from synthetic cells
Abstract
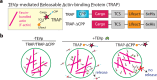
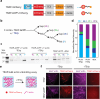
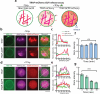
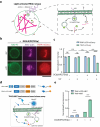
Synthetic cells offer a versatile platform for addressing biomedical and environmental challenges, due to their modular design and capability to mimic cellular processes such as biosensing, intercellular communication, and metabolism. Constructing synthetic cells capable of stimuli-responsive secretion is vital for applications in targeted drug delivery and biosensor development. Previous attempts at engineering secretion for synthetic cells have been confined to non-specific cargo release via membrane pores, limiting the spatiotemporal precision and specificity necessary for selective secretion. Here, we designed and constructed a protein-based platform termed TEV Protease-mediated Releasable Actin-binding protein (TRAP) for selective, rapid, and triggerable secretion in synthetic cells. TRAP is designed to bind tightly to reconstituted actin networks and is proteolytically released from bound actin, followed by secretion via cell-penetrating peptide membrane translocation. We demonstrated TRAP's efficacy in facilitating light-activated secretion of both fluorescent and luminescent proteins. By equipping synthetic cells with a controlled secretion mechanism, TRAP paves the way for the development of stimuli-responsive biomaterials, versatile synthetic cell-based biosensing systems, and therapeutic applications through the integration of synthetic cells with living cells for targeted delivery of protein therapeutics.
Conflict of interest statement
Conflict of Interest The authors declare no conflict of interest.
Figures
Similar articles
-
Spatiotemporal Control Over Protein Release from Artificial Cells via a Light-Activatable Protease.Adv Biol (Weinh). 2024 Sep 27:e2400353. doi: 10.1002/adbi.202400353. Online ahead of print. Adv Biol (Weinh). 2024. PMID: 39334525
-
Light-Activated Assembly of Connexon Nanopores in Synthetic Cells.J Am Chem Soc. 2023 Feb 15;145(6):3561-3568. doi: 10.1021/jacs.2c12491. Epub 2023 Feb 1. J Am Chem Soc. 2023. PMID: 36724060 Free PMC article.
-
Intelligent "Peptide-Gathering Mechanical Arm" Tames Wild "Trojan-Horse" Peptides for the Controlled Delivery of Cancer Nanotherapeutics.ACS Appl Mater Interfaces. 2017 Dec 6;9(48):41767-41781. doi: 10.1021/acsami.7b15523. Epub 2017 Nov 21. ACS Appl Mater Interfaces. 2017. PMID: 29161013
-
Functional DNA Molecules Enable Selective and Stimuli-Responsive Nanoparticles for Biomedical Applications.Acc Chem Res. 2019 Sep 17;52(9):2415-2426. doi: 10.1021/acs.accounts.9b00167. Epub 2019 Aug 14. Acc Chem Res. 2019. PMID: 31411853 Free PMC article. Review.
-
Boolean logic in synthetic biology and biomaterials: Towards living materials in mammalian cell therapeutics.Clin Transl Med. 2023 Jul;13(7):e1244. doi: 10.1002/ctm2.1244. Clin Transl Med. 2023. PMID: 37386762 Free PMC article. Review.
References
-
- Boyd M. A., Kamat N. P., Trends in Biotechnology 2021, 39, 927. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
