This is a preprint.
Adaptive protein synthesis in genetic models of copper deficiency and childhood neurodegeneration
- PMID: 39314281
- PMCID: PMC11419079
- DOI: 10.1101/2024.09.09.612106
Adaptive protein synthesis in genetic models of copper deficiency and childhood neurodegeneration
Abstract
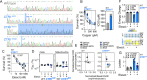
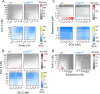
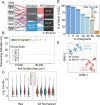
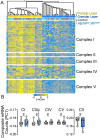
Rare inherited diseases caused by mutations in the copper transporters SLC31A1 (CTR1) or ATP7A induce copper deficiency in the brain, causing seizures and neurodegeneration in infancy through poorly understood mechanisms. Here, we used multiple model systems to characterize the molecular mechanisms by which neuronal cells respond to copper deficiency. Targeted deletion of CTR1 in neuroblastoma cells produced copper deficiency that was associated with a metabolic shift favoring glycolysis over oxidative phosphorylation. Proteomic and transcriptomic analysis of CTR1 KO cells revealed simultaneous upregulation of mTORC1 and S6K signaling and reduced PERK signaling. Patterns of gene and protein expression and pharmacogenomics show increased activation of the mTORC1-S6K pathway as a pro-survival mechanism, ultimately resulting in increased protein synthesis. Spatial transcriptomic profiling of Atp7a flx/Y :: Vil1 Cre/+ mice identified upregulated protein synthesis machinery and mTORC1-S6K pathway genes in copper-deficient Purkinje neurons in the cerebellum. Genetic epistasis experiments in Drosophila demonstrated that copper deficiency dendritic phenotypes in class IV neurons are partially rescued by increased S6k expression or 4E-BP1 (Thor) RNAi, while epidermis phenotypes are exacerbated by Akt, S6k, or raptor RNAi. Overall, we demonstrate that increased mTORC1-S6K pathway activation and protein synthesis is an adaptive mechanism by which neuronal cells respond to copper deficiency.
Conflict of interest statement
Conflict of Interest Statement The authors declare no competing financial interests.
Figures












Similar articles
-
Depressing time: Waiting, melancholia, and the psychoanalytic practice of care.In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. In: Kirtsoglou E, Simpson B, editors. The Time of Anthropology: Studies of Contemporary Chronopolitics. Abingdon: Routledge; 2020. Chapter 5. PMID: 36137063 Free Books & Documents. Review.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
Enabling Systemic Identification and Functionality Profiling for Cdc42 Homeostatic Modulators.bioRxiv [Preprint]. 2024 Jan 8:2024.01.05.574351. doi: 10.1101/2024.01.05.574351. bioRxiv. 2024. Update in: Commun Chem. 2024 Nov 19;7(1):271. doi: 10.1038/s42004-024-01352-7 PMID: 38260445 Free PMC article. Updated. Preprint.
-
Qualitative evidence synthesis informing our understanding of people's perceptions and experiences of targeted digital communication.Cochrane Database Syst Rev. 2019 Oct 23;10(10):ED000141. doi: 10.1002/14651858.ED000141. Cochrane Database Syst Rev. 2019. PMID: 31643081 Free PMC article.
-
Pharmacological treatments in panic disorder in adults: a network meta-analysis.Cochrane Database Syst Rev. 2023 Nov 28;11(11):CD012729. doi: 10.1002/14651858.CD012729.pub3. Cochrane Database Syst Rev. 2023. PMID: 38014714 Free PMC article. Review.
References
-
- Almeida LM, Pinho BR, Duchen MR, Oliveira JMA (2022) The PERKs of mitochondria protection during stress: insights for PERK modulation in neurodegenerative and metabolic diseases. Biol Rev Camb Philos Soc 97:1737–1748. - PubMed
-
- Arshadi C, Günther U, Eddison M, Harrington KIS, Ferreira TA (2021) SNT: a unifying toolbox for quantification of neuronal anatomy. Nat Methods 18:374–377. - PubMed
-
- Atkinson DE, Walton GM (1967) Adenosine triphosphate conservation in metabolic regulation. Rat liver citrate cleavage enzyme. J Biol Chem 242:3239–3241. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
