Tumor-associated mesenchymal stromal cells modulate macrophage phagocytosis in stromal-rich colorectal cancer via PD-1 signaling
- PMID: 39310770
- PMCID: PMC11416555
- DOI: 10.1016/j.isci.2024.110701
Tumor-associated mesenchymal stromal cells modulate macrophage phagocytosis in stromal-rich colorectal cancer via PD-1 signaling
Abstract
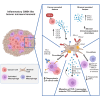
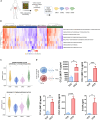
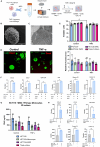
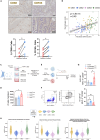
CMS4 colorectal cancer (CRC), based on the consensus molecular subtype (CMS), stratifies patients with the poorest disease-free survival rates. It is characterized by a strong mesenchymal stromal cell (MSC) signature, wound healing-like inflammation and therapy resistance. We utilized 2D and 3D in vitro, in vivo, and ex vivo models to assess the impact of inflammation and stromal cells on immunosuppression in CMS4 CRC. RNA sequencing data from untreated stage II/III CRC patients showed enriched TNF-α signatures in CMS1 and CMS4 tumors. Secretome from TNF-α treated cancer cells induced an immunomodulatory and chemotactic phenotype in MSC and cancer-associated fibroblasts (CAFs). Macrophages in CRC tumours migrate and preferentially localise in stromal compartment. Inflammatory CRC secretome enhances expression of PD-L1 and CD47 on both human and murine stromal cells. We demonstrate that TNF-α-induced inflammation in CRC suppresses macrophage phagocytosis via stromal cells. We show that stromal cell-mediated suppression of macrophage phagocytosis is mediated in part through PD-1 signaling. These data suggest that re-stratification of CRC by CMS may reveal patient subsets with microsatellite stable tumors, particularly CMS4-like tumors, that may respond to immunotherapies.
Keywords: Cancer; Immunology; Transcriptomics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no conflicts of interest.
Figures





Similar articles
-
Intratumor heterogeneity and cell secretome promote chemotherapy resistance and progression of colorectal cancer.Cell Death Dis. 2023 May 5;14(5):306. doi: 10.1038/s41419-023-05806-z. Cell Death Dis. 2023. PMID: 37142595 Free PMC article.
-
Refining colorectal cancer classification and clinical stratification through a single-cell atlas.Genome Biol. 2022 May 11;23(1):113. doi: 10.1186/s13059-022-02677-z. Genome Biol. 2022. PMID: 35538548 Free PMC article.
-
An integrative gene expression signature analysis identifies CMS4 KRAS-mutated colorectal cancers sensitive to combined MEK and SRC targeted therapy.BMC Cancer. 2022 Mar 10;22(1):256. doi: 10.1186/s12885-022-09344-3. BMC Cancer. 2022. PMID: 35272617 Free PMC article.
-
The potential role of platelets in the consensus molecular subtypes of colorectal cancer.Cancer Metastasis Rev. 2017 Jun;36(2):273-288. doi: 10.1007/s10555-017-9678-9. Cancer Metastasis Rev. 2017. PMID: 28681242 Review.
-
Investigation of colorectal cancer in accordance with consensus molecular subtype classification.Ann Gastroenterol Surg. 2020 Jul 21;4(5):528-539. doi: 10.1002/ags3.12362. eCollection 2020 Sep. Ann Gastroenterol Surg. 2020. PMID: 33005848 Free PMC article. Review.
References
-
- Källberg J., Harrison A., March V., Berzina S., Nemazanyy I., Kepp O., Kroemer G., Mouillet-Richard S., Laurent-Puig P., Taly V., Xiao W. Intratumor heterogeneity and cell secretome promote chemotherapy resistance and progression of colorectal cancer. Cell Death Dis. 2023;14:306. doi: 10.1038/s41419-023-05806-z. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

