Spatial colocalization and combined survival benefit of natural killer and CD8 T cells despite profound MHC class I loss in non-small cell lung cancer
- PMID: 39299754
- PMCID: PMC11418484
- DOI: 10.1136/jitc-2024-009126
Spatial colocalization and combined survival benefit of natural killer and CD8 T cells despite profound MHC class I loss in non-small cell lung cancer
Abstract
Background: Major histocompatibility complex class I (MHC-I) loss is frequent in non-small cell lung cancer (NSCLC) rendering tumor cells resistant to T cell lysis. NK cells kill MHC-I-deficient tumor cells, and although previous work indicated their presence at NSCLC margins, they were functionally impaired. Within, we evaluated whether NK cell and CD8 T cell infiltration and activation vary with MHC-I expression.
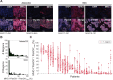
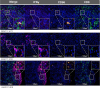
Methods: We used single-stain immunohistochemistry (IHC) and Kaplan-Meier analysis to test the effect of NK cell and CD8 T cell infiltration on overall and disease-free survival. To delineate immune covariates of MHC-I-disparate lung cancers, we used multiplexed immunofluorescence (mIF) imaging followed by multivariate statistical modeling. To identify differences in infiltration and intercellular communication between IFNγ-activated and non-activated lymphocytes, we developed a computational pipeline to enumerate single-cell neighborhoods from mIF images followed by multivariate discriminant analysis.
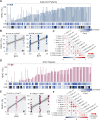
Results: Spatial quantitation of tumor cell MHC-I expression revealed intratumoral and intertumoral heterogeneity, which was associated with the local lymphocyte landscape. IHC analysis revealed that high CD56+ cell numbers in patient tumors were positively associated with disease-free survival (HR=0.58, p=0.064) and overall survival (OS) (HR=0.496, p=0.041). The OS association strengthened with high counts of both CD56+ and CD8+ cells (HR=0.199, p<1×10-3). mIF imaging and multivariate discriminant analysis revealed enrichment of both CD3+CD8+ T cells and CD3-CD56+ NK cells in MHC-I-bearing tumors (p<0.05). To infer associations of functional cell states and local cell-cell communication, we analyzed spatial single-cell neighborhood profiles to delineate the cellular environments of IFNγ+/- NK cells and T cells. We discovered that both IFNγ+ NK and CD8 T cells were more frequently associated with other IFNγ+ lymphocytes in comparison to IFNγ- NK cells and CD8 T cells (p<1×10-30). Moreover, IFNγ+ lymphocytes were most often found clustered near MHC-I+ tumor cells.
Conclusions: Tumor-infiltrating NK cells and CD8 T cells jointly affected control of NSCLC tumor progression. Coassociation of NK and CD8 T cells was most evident in MHC-I-bearing tumors, especially in the presence of IFNγ. Frequent colocalization of IFNγ+ NK cells with other IFNγ+ lymphocytes in near-neighbor analysis suggests NSCLC lymphocyte activation is coordinately regulated.
Keywords: Lung Cancer; Major histocompatibility complex - MHC; Natural killer - NK; T cell; Tumor infiltrating lymphocyte - TIL.
© Author(s) (or their employer(s)) 2024. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: CLS: Research support to the University of Virginia from Celldex (funding, drug), Merck (funding, drug), Theraclion (device staff support); Funding to the University of Virginia from Polynoma for PI role on the MAVIS Clinical Trial; Funding to the University of Virginia for roles on Scientific Advisory Boards for Immatics and CureVac. CLS also receives licensing fee payments through the UVA Licensing and Ventures Group for patents for peptides used in cancer vaccines. RDG: Research support to the University of Virginia from Pfizer, Amgen, Chugai, Merck, AstraZeneca, Janssen, Daiichi Sankyo, Alliance Foundation, Takeda, ECOG/ACRIN, Jounce Therapeutics, Bristol Myers Squibb, SWOG, Helsinn, Dizal Pharmaceuticals, and Mirati. RDG received payment for service on Scientific Advisory Boards including AstraZeneca, Takeda, Gilead, Janssen, Mirati, Daiichi Sankyo, Sanofi, Oncocyte, Jazz Pharmaceuticals, Blueprint Medicines, and Merus.
Figures



Update of
-
Spatial colocalization and combined survival benefit of natural killer and CD8 T cells despite profound MHC class I loss in non-small cell lung cancer.bioRxiv [Preprint]. 2024 Jun 25:2024.02.20.581048. doi: 10.1101/2024.02.20.581048. bioRxiv. 2024. Update in: J Immunother Cancer. 2024 Sep 18;12(9):e009126. doi: 10.1136/jitc-2024-009126 PMID: 38979183 Free PMC article. Updated. Preprint.
Similar articles
-
Spatial colocalization and combined survival benefit of natural killer and CD8 T cells despite profound MHC class I loss in non-small cell lung cancer.bioRxiv [Preprint]. 2024 Jun 25:2024.02.20.581048. doi: 10.1101/2024.02.20.581048. bioRxiv. 2024. Update in: J Immunother Cancer. 2024 Sep 18;12(9):e009126. doi: 10.1136/jitc-2024-009126 PMID: 38979183 Free PMC article. Updated. Preprint.
-
Natural killer cells infiltrating colorectal cancer and MHC class I expression.Mol Immunol. 2005 Feb;42(4):541-6. doi: 10.1016/j.molimm.2004.07.039. Mol Immunol. 2005. PMID: 15607811
-
The positive prognostic effect of stromal CD8+ tumor-infiltrating T cells is restrained by the expression of HLA-E in non-small cell lung carcinoma.Oncotarget. 2016 Jan 19;7(3):3477-88. doi: 10.18632/oncotarget.6506. Oncotarget. 2016. PMID: 26658106 Free PMC article.
-
Tumor mutational load, CD8+ T cells, expression of PD-L1 and HLA class I to guide immunotherapy decisions in NSCLC patients.Cancer Immunol Immunother. 2020 May;69(5):771-777. doi: 10.1007/s00262-020-02506-x. Epub 2020 Feb 12. Cancer Immunol Immunother. 2020. PMID: 32047958 Free PMC article.
-
Structure and function of major histocompatibility complex class I antigens.Curr Opin Organ Transplant. 2010 Aug;15(4):499-504. doi: 10.1097/MOT.0b013e32833bfb33. Curr Opin Organ Transplant. 2010. PMID: 20613521 Free PMC article. Review.
Cited by
-
Single intravenous administration of oncolytic adenovirus TILT-123 results in systemic tumor transduction and immune response in patients with advanced solid tumors.J Exp Clin Cancer Res. 2024 Nov 6;43(1):297. doi: 10.1186/s13046-024-03219-0. J Exp Clin Cancer Res. 2024. PMID: 39506856 Free PMC article. Clinical Trial.
References
-
- Travis WD, Brambilla E, Noguchi M, et al. International Association for the Study of Lung Cancer/American Thoracic Society/European Respiratory Society: international multidisciplinary classification of lung adenocarcinoma: executive summary. Proc Am Thorac Soc. 2011;8:381–5. doi: 10.1513/pats.201107-042ST. - DOI - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous
