HLA A*24:02-restricted T cell receptors cross-recognize bacterial and preproinsulin peptides in type 1 diabetes
- PMID: 39286976
- PMCID: PMC11405051
- DOI: 10.1172/JCI164535
HLA A*24:02-restricted T cell receptors cross-recognize bacterial and preproinsulin peptides in type 1 diabetes
Abstract
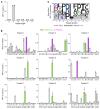
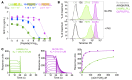
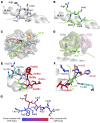
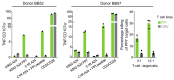
CD8+ T cells destroy insulin-producing pancreatic β cells in type 1 diabetes through HLA class I-restricted presentation of self-antigens. Combinatorial peptide library screening was used to produce a preferred peptide recognition landscape for a patient-derived T cell receptor (TCR) that recognized the preproinsulin-derived (PPI-derived) peptide sequence LWMRLLPLL in the context of disease risk allele HLA A*24:02. Data were used to generate a strong superagonist peptide, enabling production of an autoimmune HLA A*24:02-peptide-TCR structure by crystal seeding. TCR binding to the PPI epitope was strongly focused on peptide residues Arg4 and Leu5, with more flexibility at other positions, allowing the TCR to strongly engage many peptides derived from pathogenic bacteria. We confirmed an epitope from Klebsiella that was recognized by PPI-reactive T cells from 3 of 3 HLA A*24:02+ patients. Remarkably, the same epitope selected T cells from 7 of 8 HLA A*24+ healthy donors that cross-reacted with PPI, leading to recognition and killing of HLA A*24:02+ cells expressing PPI. These data provide a mechanism by which molecular mimicry between pathogen and self-antigens could have resulted in the breaking of self-tolerance to initiate disease.
Keywords: Autoimmunity; Diabetes; Immunology; Structural biology; T cell receptor.
Figures



Comment in
- Microbial mimics supersize the pathogenic self-response
Similar articles
-
Circulating preproinsulin signal peptide-specific CD8 T cells restricted by the susceptibility molecule HLA-A24 are expanded at onset of type 1 diabetes and kill β-cells.Diabetes. 2012 Jul;61(7):1752-9. doi: 10.2337/db11-1520. Epub 2012 Apr 20. Diabetes. 2012. PMID: 22522618 Free PMC article.
-
CD8 T cell autoreactivity to preproinsulin epitopes with very low human leucocyte antigen class I binding affinity.Clin Exp Immunol. 2012 Oct;170(1):57-65. doi: 10.1111/j.1365-2249.2012.04635.x. Clin Exp Immunol. 2012. PMID: 22943201 Free PMC article.
-
Circulating β cell-specific CD8+ T cells restricted by high-risk HLA class I molecules show antigen experience in children with and at risk of type 1 diabetes.Clin Exp Immunol. 2020 Mar;199(3):263-277. doi: 10.1111/cei.13391. Epub 2019 Nov 10. Clin Exp Immunol. 2020. PMID: 31660582 Free PMC article.
-
T cell recognition of autoantigens in human type 1 diabetes: clinical perspectives.Clin Dev Immunol. 2011;2011:513210. doi: 10.1155/2011/513210. Epub 2011 Jul 19. Clin Dev Immunol. 2011. PMID: 21785617 Free PMC article. Review.
-
Immunogenic self-peptides - the great unknowns in autoimmunity: Identifying T-cell epitopes driving the autoimmune response in autoimmune diseases.Front Immunol. 2023 Jan 9;13:1097871. doi: 10.3389/fimmu.2022.1097871. eCollection 2022. Front Immunol. 2023. PMID: 36700227 Free PMC article. Review.
References
-
- Foulis AK, et al. Aberrant expression of class II major histocompatibility complex molecules by B cells and hyperexpression of class I major histocompatibility complex molecules by insulin containing islets in type 1 (insulin-dependent) diabetes mellitus. Diabetologia. 1987;30(5):333–343. doi: 10.1007/BF00299027. - DOI - PMC - PubMed
-
- Itoh N, et al. Mononuclear cell infiltration and its relation to the expression of major histocompatibility complex antigens and adhesion molecules in pancreas biopsy specimens from newly diagnosed insulin-dependent diabetes mellitus patients. J Clin Invest. 1993;92(5):2313–2322. doi: 10.1172/JCI116835. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

