PRDM3/16 regulate chromatin accessibility required for NKX2-1 mediated alveolar epithelial differentiation and function
- PMID: 39284798
- PMCID: PMC11405758
- DOI: 10.1038/s41467-024-52154-3
PRDM3/16 regulate chromatin accessibility required for NKX2-1 mediated alveolar epithelial differentiation and function
Abstract
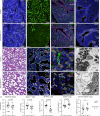
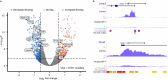
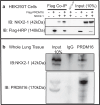
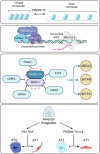
While the critical role of NKX2-1 and its transcriptional targets in lung morphogenesis and pulmonary epithelial cell differentiation is increasingly known, mechanisms by which chromatin accessibility alters the epigenetic landscape and how NKX2-1 interacts with other co-activators required for alveolar epithelial cell differentiation and function are not well understood. Combined deletion of the histone methyl transferases Prdm3 and Prdm16 in early lung endoderm causes perinatal lethality due to respiratory failure from loss of AT2 cells and the accumulation of partially differentiated AT1 cells. Combination of single-cell RNA-seq, bulk ATAC-seq, and CUT&RUN data demonstrate that PRDM3 and PRDM16 regulate chromatin accessibility at NKX2-1 transcriptional targets critical for perinatal AT2 cell differentiation and surfactant homeostasis. Lineage specific deletion of PRDM3/16 in AT2 cells leads to lineage infidelity, with PRDM3/16 null cells acquiring partial AT1 fate. Together, these data demonstrate that NKX2-1-dependent regulation of alveolar epithelial cell differentiation is mediated by epigenomic modulation via PRDM3/16.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Update of
-
PRDM3/16 Regulate Chromatin Accessibility Required for NKX2-1 Mediated Alveolar Epithelial Differentiation and Function.bioRxiv [Preprint]. 2023 Dec 20:2023.12.20.570481. doi: 10.1101/2023.12.20.570481. bioRxiv. 2023. Update in: Nat Commun. 2024 Sep 16;15(1):8112. doi: 10.1038/s41467-024-52154-3. PMID: 38187557 Free PMC article. Updated. Preprint.
Similar articles
-
PRDM3/16 Regulate Chromatin Accessibility Required for NKX2-1 Mediated Alveolar Epithelial Differentiation and Function.bioRxiv [Preprint]. 2023 Dec 20:2023.12.20.570481. doi: 10.1101/2023.12.20.570481. bioRxiv. 2023. Update in: Nat Commun. 2024 Sep 16;15(1):8112. doi: 10.1038/s41467-024-52154-3. PMID: 38187557 Free PMC article. Updated. Preprint.
-
Differential chromatin binding of the lung lineage transcription factor NKX2-1 resolves opposing murine alveolar cell fates in vivo.Nat Commun. 2021 May 4;12(1):2509. doi: 10.1038/s41467-021-22817-6. Nat Commun. 2021. PMID: 33947861 Free PMC article.
-
Transcriptional control of lung alveolar type 1 cell development and maintenance by NK homeobox 2-1.Proc Natl Acad Sci U S A. 2019 Oct 8;116(41):20545-20555. doi: 10.1073/pnas.1906663116. Epub 2019 Sep 23. Proc Natl Acad Sci U S A. 2019. PMID: 31548395 Free PMC article.
-
Emerging Roles of PRDM Factors in Stem Cells and Neuronal System: Cofactor Dependent Regulation of PRDM3/16 and FOG1/2 (Novel PRDM Factors).Cells. 2020 Dec 4;9(12):2603. doi: 10.3390/cells9122603. Cells. 2020. PMID: 33291744 Free PMC article. Review.
-
Commitment and differentiation of lung cell lineages.Biochem Cell Biol. 1998;76(6):971-95. Biochem Cell Biol. 1998. PMID: 10392710 Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

