HCMV detection in Asian gastric cancer RNA-seq data sets and clinical validation in Indian GC patients reveals the HCMV-GC specific gene signatures
- PMID: 39283078
- PMCID: PMC11494955
- DOI: 10.1128/msystems.00673-24
HCMV detection in Asian gastric cancer RNA-seq data sets and clinical validation in Indian GC patients reveals the HCMV-GC specific gene signatures
Abstract
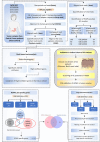
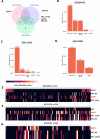
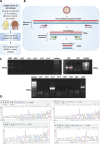
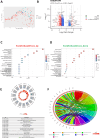
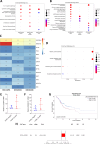
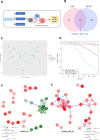
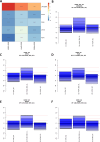
Gastric cancer (GC) prevalence is very high in the Asian population. Oncogenic viruses play a crucial role in the progression of different types of cancers. Through reanalysis of clinical RNA-seq data sets derived from Asian GC patients, this study identified the presence of human cytomegalovirus (HCMV) in Asian GC tumors, next to the well-studied association of EBV. Clinical recruitment of the Indian GC cohort and screening for HCMV presence identified a 14.28% occurrence, similar to that observed in the bioinformatics analysis. A combinatorial approach of rank-based meta-analysis and ranking of groups based on an expectation-maximization algorithm identified that the upregulated LINC02864 and MAGEA10 correlated with poor survival of GC patients and downregulated tumor suppressor genes enriching for gastric acid secretion pathway to be associated with HCMV-positive GC patients, revealing the progressive role of HCMV infection in GC. Genes that discriminate between different stages of GC were identified through feature selection implemented in a machine-learning approach. LTF and KLK10 expressions were found to be specifically dysregulated by HCMV and can also indicate the GC stages. The results of this study will guide future studies to identify the functional role of these genes in the HCMV-associated GC.IMPORTANCENearly 75% of gastric cancer (GC) cases reported globally are from the Asian population. Most existing public databases, such as TCGA, comprise only a fractional portion of data derived from Asian ancestry. This study identified EBV and human cytomegalovirus (HCMV)'s higher detection in GC patients. The presence and role of EBV associated with GC are well-known, and the observation of HCMV prompted us to validate the findings in a small cohort of 40 Indian GC patients. We observed a 14.28% occurrence of HCMV in the Indian cohort, similar to that observed from next-generation sequencing. A combinatorial approach of rank-based meta-analysis and ranking of groups based on an expectation-maximization algorithm identified that the upregulated LINC02864 and MAGEA10 correlated with poor survival of GC patients and downregulated tumor suppressor genes enriching for gastric acid secretion pathway to be associated with HCMV-positive GC patients, revealing the progressive role of HCMV infection in GC.
Keywords: HCMV; RNA-seq; clinical screening; gastric cancer; meta-analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Human cytomegalovirus detection in gastric cancer and its possible association with lymphatic metastasis.Diagn Microbiol Infect Dis. 2017 May;88(1):62-68. doi: 10.1016/j.diagmicrobio.2017.02.001. Epub 2017 Feb 8. Diagn Microbiol Infect Dis. 2017. PMID: 28238538
-
Human cytomegalovirus latent infection alters the expression of cellular and viral microRNA.Gene. 2014 Feb 25;536(2):272-8. doi: 10.1016/j.gene.2013.12.012. Epub 2013 Dec 18. Gene. 2014. PMID: 24361963
-
Comprehensive bioinformatics analysis of human cytomegalovirus pathway genes in pan-cancer.Hum Genomics. 2024 Jun 17;18(1):65. doi: 10.1186/s40246-024-00633-5. Hum Genomics. 2024. PMID: 38886862 Free PMC article.
-
Potential prognostic impact of EBV RNA-seq reads in gastric cancer: a reanalysis of The Cancer Genome Atlas cohort.FEBS Open Bio. 2020 Mar;10(3):455-467. doi: 10.1002/2211-5463.12803. Epub 2020 Feb 16. FEBS Open Bio. 2020. PMID: 31991047 Free PMC article.
-
Cytomegalovirus Infection Is a Risk Factor in Gastrointestinal Cancer: A Cross-Sectional and Meta-Analysis Study.Intervirology. 2020;63(1-6):10-16. doi: 10.1159/000506683. Epub 2020 Aug 7. Intervirology. 2020. PMID: 32772018 Review.
References
-
- Wang H, Chen X-L, Liu K, Bai D, Zhang W-H, Chen X-Z, Hu J-K, SIGES research group . 2020. Associations between gastric cancer risk and virus infection other than Epstein-Barr virus: a systematic review and meta-analysis based on epidemiological studies. Clin Transl Gastroenterol 11:e00201. doi:10.14309/ctg.0000000000000201 - DOI - PMC - PubMed
MeSH terms
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
