This is a preprint.
SARS-CoV-2 Nsp15 antagonizes the cGAS-STING-mediated antiviral innate immune responses
- PMID: 39282446
- PMCID: PMC11398466
- DOI: 10.1101/2024.09.05.611469
SARS-CoV-2 Nsp15 antagonizes the cGAS-STING-mediated antiviral innate immune responses
Abstract
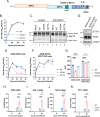
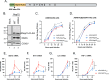
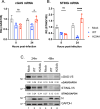
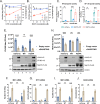
Coronavirus (CoV) Nsp15 is a viral endoribonuclease (EndoU) with a preference for uridine residues. CoV Nsp15 is an innate immune antagonist which prevents dsRNA sensor recognition and stress granule formation by targeting viral and host RNAs. SARS-CoV-2 restricts and delays the host antiviral innate immune responses through multiple viral proteins, but the role of SARS-CoV-2 Nsp15 in innate immune evasion is not completely understood. Here, we generate an EndoU activity knockout rSARS-CoV-2Nsp15-H234A to elucidate the biological functions of Nsp15. Relative to wild-type rSARS-CoV-2, replication of rSARS-CoV-2Nsp15-H234A was significantly decreased in IFN-responsive A549-ACE2 cells but not in its STAT1 knockout counterpart. Transcriptomic analysis revealed upregulation of innate immune response genes in cells infected with rSARS-CoV-2Nsp15-H234A relative to wild-type virus, including cGAS-STING, cytosolic DNA sensors activated by both DNA and RNA viruses. Treatment with STING inhibitors H-151 and SN-011 rescued the attenuated phenotype of rSARS-CoV-2Nsp15-H234A. SARS-CoV-2 Nsp15 inhibited cGAS-STING-mediated IFN-β promoter and NF-κB reporter activity, as well as facilitated the replication of EV-D68 and NDV by diminishing cGAS and STING expression and downstream innate immune responses. Notably, the decline in cGAS and STING was also apparent during SARS-CoV-2 infection. The EndoU activity was essential for SARS-CoV-2 Nsp15-mediated cGAS and STING downregulation, but not all HCoV Nsp15 share the consistent substrate selectivity. In the hamster model, rSARS-CoV-2Nsp15-H234A replicated to lower titers in the nasal turbinates and lungs and induced higher innate immune responses. Collectively, our findings exhibit that SARS-CoV-2 Nsp15 serves as a host innate immune antagonist by targeting host cGAS and STING.
Keywords: Biological Sciences/Microbiology; Nsp15; SARS-CoV-2; cGAS-STING; innate immunity.
Conflict of interest statement
Competing Interest Statement: S.J. is a co-founder of Elucidate Bio Inc, has received speaking honorariums from Cell Signaling Technology, and has received research support from Roche unrelated to this work.
Figures


Similar articles
-
SARS-CoV-2 nsp15 endoribonuclease antagonizes dsRNA-induced antiviral signaling.Proc Natl Acad Sci U S A. 2024 Apr 9;121(15):e2320194121. doi: 10.1073/pnas.2320194121. Epub 2024 Apr 3. Proc Natl Acad Sci U S A. 2024. PMID: 38568967 Free PMC article.
-
SARS-CoV-2 nsp15 endoribonuclease antagonizes dsRNA-induced antiviral signaling.bioRxiv [Preprint]. 2023 Nov 15:2023.11.15.566945. doi: 10.1101/2023.11.15.566945. bioRxiv. 2023. Update in: Proc Natl Acad Sci U S A. 2024 Apr 9;121(15):e2320194121. doi: 10.1073/pnas.2320194121. PMID: 38014074 Free PMC article. Updated. Preprint.
-
MERS-CoV endoribonuclease and accessory proteins jointly evade host innate immunity during infection of lung and nasal epithelial cells.bioRxiv [Preprint]. 2021 Dec 21:2021.12.20.473564. doi: 10.1101/2021.12.20.473564. bioRxiv. 2021. Update in: Proc Natl Acad Sci U S A. 2022 May 24;119(21):e2123208119. doi: 10.1073/pnas.2123208119. PMID: 34981054 Free PMC article. Updated. Preprint.
-
The coronavirus nsp15 endoribonuclease: A puzzling protein and pertinent antiviral drug target.Antiviral Res. 2024 Aug;228:105921. doi: 10.1016/j.antiviral.2024.105921. Epub 2024 May 31. Antiviral Res. 2024. PMID: 38825019 Review.
-
Progress of cGAS-STING signaling in response to SARS-CoV-2 infection.Front Immunol. 2022 Dec 7;13:1010911. doi: 10.3389/fimmu.2022.1010911. eCollection 2022. Front Immunol. 2022. PMID: 36569852 Free PMC article. Review.
References
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
