Network medicine based approach for identifying the type 2 diabetes, osteoarthritis and triple negative breast cancer interactome: Finding the hub of hub genes
- PMID: 39281650
- PMCID: PMC11401126
- DOI: 10.1016/j.heliyon.2024.e36650
Network medicine based approach for identifying the type 2 diabetes, osteoarthritis and triple negative breast cancer interactome: Finding the hub of hub genes
Abstract
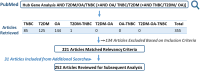
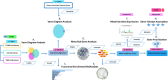
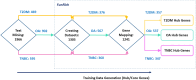
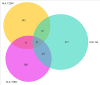
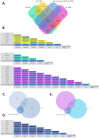
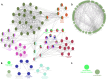
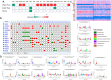
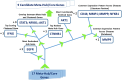
The increasing prevalence of multi-morbidities, particularly the incidence of breast cancer in diabetic/osteoarthritic patients emphasize on the need for exploring the underlying molecular mechanisms resulting in carcinogenesis. To address this, present study employed a systems biology approach to identify switch genes pivotal to the crosstalk between diseased states resulting in multi-morbid conditions. Hub genes previously reported for type 2 diabetes mellitus (T2DM), osteoarthritis (OA), and triple negative breast cancer (TNBC), were extracted from published literature and fed into an integrated bioinformatics analyses pipeline. Thirty-one hub genes common to all three diseases were identified. Functional enrichment analyses showed these were mainly enriched for immune and metabolism associated terms including advanced glycation end products (AGE) pathways, cancer pathways, particularly breast neoplasm, immune system signalling and adipose tissue. The T2DM-OA-TNBC interactome was subjected to protein-protein interaction network analyses to identify meta hub/clustered genes. These were prioritized and wired into a three disease signalling map presenting the enriched molecular crosstalk on T2DM-OA-TNBC axes to gain insight into the molecular mechanisms underlying disease-disease interactions. Deciphering the molecular bases for the intertwined metabolic and immune states may potentiate the discovery of biomarkers critical for identifying and targeting the immuno-metabolic origin of disease.
Keywords: Bioinformatics; Gene mining; Meta hub genes; Network analysis; Osteoarthritis; Switch genes; Triple negative breast cancer; Type 2 diabetes mellitus.
© 2024 The Authors. Published by Elsevier Ltd.
Conflict of interest statement
None Declared.
Figures



Similar articles
-
Novel biomarkers identified in triple-negative breast cancer through RNA-sequencing.Clin Chim Acta. 2022 Jun 1;531:302-308. doi: 10.1016/j.cca.2022.04.990. Epub 2022 Apr 30. Clin Chim Acta. 2022. PMID: 35504321
-
Identification of a five genes prognosis signature for triple-negative breast cancer using multi-omics methods and bioinformatics analysis.Cancer Gene Ther. 2022 Nov;29(11):1578-1589. doi: 10.1038/s41417-022-00473-2. Epub 2022 Apr 26. Cancer Gene Ther. 2022. PMID: 35474355
-
Identification of Hub Genes and Pathways of Triple Negative Breast Cancer by Expression Profiles Analysis.Cancer Manag Res. 2021 Mar 1;13:2095-2104. doi: 10.2147/CMAR.S295951. eCollection 2021. Cancer Manag Res. 2021. PMID: 33688252 Free PMC article.
-
Identification of miRNAs and related hub genes associated with the triple negative breast cancer using integrated bioinformatics analysis and in vitro approach.J Biomol Struct Dyn. 2022;40(22):11676-11690. doi: 10.1080/07391102.2021.1961869. Epub 2021 Aug 13. J Biomol Struct Dyn. 2022. PMID: 34387138
-
Identification of Disease-Specific Hub Biomarkers and Immune Infiltration in Osteoarthritis and Rheumatoid Arthritis Synovial Tissues by Bioinformatics Analysis.Dis Markers. 2021 May 17;2021:9911184. doi: 10.1155/2021/9911184. eCollection 2021. Dis Markers. 2021. PMID: 34113405 Free PMC article.
References
-
- Grimaldi A.M., Conte F., Pane K., Fiscon G., Mirabelli P., Baselice S., Giannatiempo R., Messina F., Franzese M., Salvatore M., Paci P., Incoronato M. The new Paradigm of network medicine to analyze breast cancer phenotypes. Int. J. Mol. Sci. 2020;21:6690. doi: 10.3390/ijms21186690. - DOI - PMC - PubMed
-
- Feodoroff M., Harjutsalo V., Mäkimattila S., Groop P.-H. Incidence and risk factors for cancer in people with type 1 diabetes, stratified by stages of diabetic kidney disease: a nationwide Finnish cohort study. The Lancet Regional Health – Europe. 2024;40 doi: 10.1016/j.lanepe.2024.100884. - DOI - PMC - PubMed
Further reading
-
- Rosshirt N., Hagmann S., Tripel E., Gotterbarm T., Kirsch J., Zeifang F., Lorenz H.‐M., Tretter T., Moradi B. A predominant Th1 polarization is present in synovial fluid of end‐stage osteoarthritic knee joints: analysis of peripheral blood, synovial fluid and synovial membrane. Clin. Exp. Immunol. 2019;195:395–406. doi: 10.1111/cei.13230. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

