Unraveling the Mystery of Energy-Sensing Enzymes and Signaling Pathways in Tumorigenesis and Their Potential as Therapeutic Targets for Cancer
- PMID: 39273044
- PMCID: PMC11394487
- DOI: 10.3390/cells13171474
Unraveling the Mystery of Energy-Sensing Enzymes and Signaling Pathways in Tumorigenesis and Their Potential as Therapeutic Targets for Cancer
Abstract
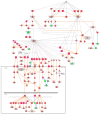
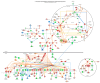
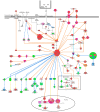
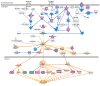
Cancer research has advanced tremendously with the identification of causative genes, proteins, and signaling pathways. Numerous antitumor drugs have been designed and screened for cancer therapeutics; however, designing target-specific drugs for malignant cells with minimal side effects is challenging. Recently, energy-sensing- and homeostasis-associated molecules and signaling pathways playing a role in proliferation, apoptosis, autophagy, and angiogenesis have received increasing attention. Energy-metabolism-based studies have shown the contribution of energetics to cancer development, where tumor cells show increased glycolytic activity and decreased oxidative phosphorylation (the Warburg effect) in order to obtain the required additional energy for rapid division. The role of energy homeostasis in the survival of normal as well as malignant cells is critical; therefore, fuel intake and expenditure must be balanced within acceptable limits. Thus, energy-sensing enzymes detecting the disruption of glycolysis, AMP, ATP, or GTP levels are promising anticancer therapeutic targets. Here, we review the common energy mediators and energy sensors and their metabolic properties, mechanisms, and associated signaling pathways involved in carcinogenesis, and explore the possibility of identifying drugs for inhibiting the energy metabolism of tumor cells. Furthermore, to corroborate our hypothesis, we performed meta-analysis based on transcriptomic profiling to search for energy-associated biomarkers and canonical pathways.
Keywords: Warburg effect; anticancer drug; cancer; energy-sensing enzymes; genes and signaling pathways.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures


Similar articles
-
Dysregulation of metabolic enzymes in tumor and stromal cells: Role in oncogenesis and therapeutic opportunities.Cancer Lett. 2020 Mar 31;473:176-185. doi: 10.1016/j.canlet.2020.01.003. Epub 2020 Jan 7. Cancer Lett. 2020. PMID: 31923436 Free PMC article. Review.
-
Cancer Metabolism: Phenotype, Signaling and Therapeutic Targets.Cells. 2020 Oct 16;9(10):2308. doi: 10.3390/cells9102308. Cells. 2020. PMID: 33081387 Free PMC article. Review.
-
Non-metabolic functions of glycolytic enzymes in tumorigenesis.Oncogene. 2017 May 11;36(19):2629-2636. doi: 10.1038/onc.2016.410. Epub 2016 Oct 31. Oncogene. 2017. PMID: 27797379 Review.
-
[The Warburg effect: from theory to therapeutic applications in cancer].Med Sci (Paris). 2013 Nov;29(11):1026-33. doi: 10.1051/medsci/20132911020. Epub 2013 Nov 20. Med Sci (Paris). 2013. PMID: 24280507 Review. French.
-
18β-glycyrrhetinic acid inhibited mitochondrial energy metabolism and gastric carcinogenesis through methylation-regulated TLR2 signaling pathway.Carcinogenesis. 2019 Apr 29;40(2):234-245. doi: 10.1093/carcin/bgy150. Carcinogenesis. 2019. PMID: 30364936
References
-
- WHO Cancer. [(accessed on 19 October 2022)]. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

