Genetic Structure and Genome-Wide Association Analysis of Growth and Reproductive Traits in Fengjing Pigs
- PMID: 39272234
- PMCID: PMC11394163
- DOI: 10.3390/ani14172449
Genetic Structure and Genome-Wide Association Analysis of Growth and Reproductive Traits in Fengjing Pigs
Abstract
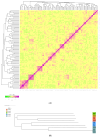
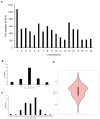
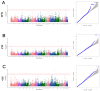
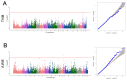
The Fengjing pig is one of the local pig breed resources in China and has many excellent germplasm characteristics. However, research on its genome is lacking. To explore the degree of genetic diversity of the Fengjing pig and to deeply explore its excellent traits, this study took Fengjing pigs as the research object and used the Beadchip Array Infinium iSelect-96|XT KPS_PorcineBreedingChipV2 for genotyping. We analyzed the genetic diversity, relatedness, inbreeding coefficient, and population structure within the Fengjing pig population. Our findings revealed that the proportion of polymorphic markers (PN) was 0.469, and the effective population size was 6.8. The observed and expected heterozygosity were 0.301 and 0.287, respectively. The G-matrix results indicated moderate relatedness within the population, with certain individuals exhibiting closer genetic relationships. The NJ evolutionary tree classified Fengjing boars into five family lines. The average inbreeding coefficient based on ROH was 0.318, indicating a high level of inbreeding. GWAS identified twenty SNPs significantly associated with growth traits (WW, 2W, and 4W) and reproductive traits (TNB and AWB). Notably, WNT8B, RAD21, and HAO1 emerged as candidate genes influencing 2W, 4W, and TNB, respectively. Genes such as WNT8B were verified by querying the PigBiobank database. In conclusion, this study provides a foundational reference for the conservation and utilization of Fengjing pig germplasm resources and offers insights for future molecular breeding efforts in Fengjing pigs.
Keywords: Fengjing pigs; SNP chip; genetic diversity; genome-wide association study.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures



Similar articles
-
Detection and Analysis of Antidiarrheal Genes and Immune Factors in Various Shanghai Pig Breeds.Biomolecules. 2024 May 17;14(5):595. doi: 10.3390/biom14050595. Biomolecules. 2024. PMID: 38786002 Free PMC article.
-
An imputation-based genome-wide association study for growth and fatness traits in Sujiang pigs.Animal. 2022 Aug;16(8):100591. doi: 10.1016/j.animal.2022.100591. Epub 2022 Jul 21. Animal. 2022. PMID: 35872387
-
Genetic diversity and population structure of Tongcheng pigs in China using whole-genome SNP chip.Front Genet. 2022 Aug 25;13:910521. doi: 10.3389/fgene.2022.910521. eCollection 2022. Front Genet. 2022. PMID: 36092902 Free PMC article.
-
Identifying Candidate Genes for Litter Size and Three Morphological Traits in Youzhou Dark Goats Based on Genome-Wide SNP Markers.Genes (Basel). 2023 May 29;14(6):1183. doi: 10.3390/genes14061183. Genes (Basel). 2023. PMID: 37372363 Free PMC article. Review.
-
Survey of SNPs Associated with Total Number Born and Total Number Born Alive in Pig.Genes (Basel). 2020 Apr 30;11(5):491. doi: 10.3390/genes11050491. Genes (Basel). 2020. PMID: 32365801 Free PMC article. Review.
References
-
- Yang J., Wu J., Ding W., Chen J., Xing J. The complete mitochondrial genome of the Fengjing pig. Mitochondrial DNA Part B. 2019;4:443–445. doi: 10.1080/23802359.2018.1555014. - DOI
-
- Chen H.-Y., Li S.-F., Gao X.-C., Li N., Liu Y.-H., Lu C.-G. Current status, conservation and exploitation of Fengjing pig resources. Shanghai Anim. Husb. Vet. Commun. 2021;1:46–47. doi: 10.14170/j.cnki.cn31-1278/s.2021.01.015. - DOI
-
- Ge Q., Gao C., Cai Y., Jiao T., Quan J., Guo Y., Zheng W., Zhao S. Evaluating genetic diversity and identifying priority conservation for seven Tibetan pig populations in China based on the mtDNA D-loop. Asian-Australas. J. Anim. Sci. 2020;33:1905–1911. doi: 10.5713/ajas.19.0752. - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

