Genetic polymorphism in untranslated regions of PRKCZ influences mRNA structure, stability and binding sites
- PMID: 39272077
- PMCID: PMC11401371
- DOI: 10.1186/s12885-024-12900-8
Genetic polymorphism in untranslated regions of PRKCZ influences mRNA structure, stability and binding sites
Abstract
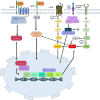
Background: Variations in untranslated regions (UTR) alter regulatory pathways impacting phenotype, disease onset, and course of disease. Protein kinase C Zeta (PRKCZ), a serine-threonine kinase, is implicated in cardiovascular, neurological and oncological disorders. Due to limited research on PRKCZ, this study aimed to investigate the impact of UTR genetic variants' on binding sites for transcription factors and miRNA. RNA secondary structure, eQTLs, and variation tolerance analysis were also part of the study.
Methods: The data related to PRKCZ gene variants was downloaded from the Ensembl genome browser, COSMIC and gnomAD. The RegulomeDB database was used to assess the functional impact of 5' UTR and 3'UTR variants. The analysis of the transcription binding sites (TFBS) was done through the Alibaba tool, and the Kyoto Encyclopaedia of Genes and Genomes (KEGG) was employed to identify pathways associated with PRKCZ. To predict the effect of variants on microRNA binding sites, PolymiRTS was utilized for 3' UTR variants, and the SNPinfo tool was used for 5' UTR variants.
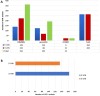
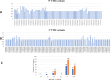
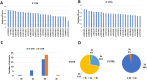
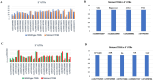
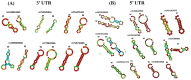
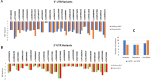
Results: The results obtained indicated that a total of 24 variants present in the 3' UTR and 25 variants present in the 5' UTR were most detrimental. TFBS analysis revealed that 5' UTR variants added YY1, repressor, and Oct1, whereas 3' UTR variants added AP-2alpha, AhR, Da, GR, and USF binding sites. The study predicted TFs that influenced PRKCZ expression. RNA secondary structure analysis showed that eight 5' UTR and six 3' UTR altered the RNA structure by either removal or addition of the stem-loop. The microRNA binding site analysis highlighted that seven 3' UTR and one 5' UTR variant altered the conserved site and also created new binding sites. eQTLs analysis showed that one variant was associated with PRKCZ expression in the lung and thyroid. The variation tolerance analysis revealed that PRKCZ was an intolerant gene.
Conclusion: This study laid the groundwork for future studies aimed at targeting PRKCZ as a therapeutic target.
Keywords: In-silico tools; Non-coding SNPs; Oncological disorders; PRKCZ; UTR; miRNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Global cataloguing of variations in untranslated regions of viral genome and prediction of key host RNA binding protein-microRNA interactions modulating genome stability in SARS-CoV-2.PLoS One. 2020 Aug 11;15(8):e0237559. doi: 10.1371/journal.pone.0237559. eCollection 2020. PLoS One. 2020. PMID: 32780783 Free PMC article.
-
PRKCE non-coding variants influence on transcription as well as translation of its gene.RNA Biol. 2022 Jan;19(1):1115-1129. doi: 10.1080/15476286.2022.2139110. RNA Biol. 2022. PMID: 36299231 Free PMC article.
-
Investigation of UTR Variants by Computational Approaches Reveal Their Functional Significance in PRKCI Gene Regulation.Genes (Basel). 2023 Jan 18;14(2):247. doi: 10.3390/genes14020247. Genes (Basel). 2023. PMID: 36833174 Free PMC article.
-
Implications of polyadenylation in health and disease.Nucleus. 2014;5(6):508-19. doi: 10.4161/nucl.36360. Epub 2014 Oct 31. Nucleus. 2014. PMID: 25484187 Free PMC article. Review.
-
A systematic analysis of disease-associated variants in the 3' regulatory regions of human protein-coding genes II: the importance of mRNA secondary structure in assessing the functionality of 3' UTR variants.Hum Genet. 2006 Oct;120(3):301-33. doi: 10.1007/s00439-006-0218-x. Epub 2006 Jun 29. Hum Genet. 2006. PMID: 16807757 Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

